code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
# Hands-on LH2: the multijunction launcher
A tokamak a intrinsequally a pulsed machine. In order to perform long plasma discharges, it is necessary to drive a part of the plasma current, in order to limit (or ideally cancel) the magnetic flux consumption. Additional current drive systems are used for these. Among the current drive systems, _Lower Hybrid Current Drive_ (LHCD) systems have demonstrated the larger current drive efficiency.
A Lower Hybrid launcher is generally made of numerous waveguides, stacked next to each other by their large sides. Since there is a phase shift between each waveguide in the toroidal direction, these launchers constitute a _phased array_. A phased array is an array of radiating elements in which the relative phases of the respective signals feeding these elements are varied in such a way that the effective radiation pattern of the array is reinforced in a desired direction and suppressed in undesired directions. A phased array is an example of N-slit diffraction. It may also be viewed as the coherent addition of N sources. In the case of LHCD launchers, the RF power is transmitted into a specific direction in the plasma through plasma waves. These waves will ultimately drive some additional plasma current in the tokamak.
The total number of waveguides in a LHCD launcher increased with each launcher generation. From simple structures, made of two or four waveguides, today's LHCD launchers such as the Tore Supra ones have hundred of waveguides. With such a number of waveguides, it is not possible anymore to excite each waveguides separately. _Multijunctions_ have been designed to solve this challenge. They act as a power splitter while phasing the adjacent waveguides.
The aim of this hands-on is to measure and characterize the RF performances of a multijunction structure, on a real Tore Supra C3 mock-up. From these relative measurements, you will have to analyse the performances of the multijunction and to deduce if the manufacturing tolerances affect them.
Before or in parallel to making measurements, you will have to calculate what would be the ideal spectral power density (or _spectrum_) launched by such a multijunction. This will serve them for making comparison theory vs experimental and thus discuss about the performances one could expect.
## 1. LHCD launcher spectrum
The objective of this section is to get you to understand the multiple requirements of a LHCD launcher.
## 2. RF Measurements
Because of the non-standard dimensions of the waveguides, it is not possible to use commercial calibration toolkit, and thus to make an
precise and absolute calibration of the RF measurements. However, relative comparisons are still relevant.
-----
# Solution
This notebook illustrates a method to compute the power density spectrum of a phased array.
```
%pylab
%matplotlib inline
from scipy.constants import c
```
## Phased array geometry
A linear phased array with equal spaced elements is easiest to analyze. It is illustrated in the following figure, in which radiating elements are located at a $\Delta z$ distance between each other. These elements radiates with a phase shift $\Delta \Phi$.
Let's a rectangular waveguide phased array facing the plasma. The waveguides periodicity is $\Delta z=b+e$ where $b$ is the width of a waveguide and $e$ the thickness of the septum between waveguides and the phase shift between waveguides is $\Delta \Phi$. We will suppose here that the amplitude and the phase of the waves in a waveguide do not depend of the spatial direction $z$. Thus we have $\Delta\Phi=$. The geometry is illustrated in the figure below.
<img src="./LH2_Multijunction_data/phased_array_grill.png">
## Ideal grill
Let's make the hypothesis that the electric field at the interface antenna-plasma is not perturbed from the electric field in the waveguide. At the antenna-plasma interface, the total electric field is thus:
$$ E(z) = \sum_{n=1}^N E_n \Pi_n(z) = \sum_{n=1}^N A_n e^{j\Phi_n} \Pi_n(z) $$
where $\Pi_n(z)$ is a Heavisyde step function, equals to unity for $z$ corresponding to a waveguide $n$, and zero elsewhere. The power density spectrum is homogeneous to the square of the Fourier transform of the electric field, that is, to :
$$ \tilde{E}(k_z) = \int E(z) e^{j k_z z} \, dz $$
Where $k_z=n_z k_0$. If one calculates the modulus square of the previous expression, this should give:
$$ dP(n_z) \propto sinc^2 \left( k_0 n_z \frac{b}{2} \right) \left( \frac{\sin(N \Phi/2)}{\sin\Phi/2} \right)^2 $$
With $\Phi=k_0 n_z \Delta z + \Delta\Phi$. The previous expression is maximized for $\Phi=2 p \pi, p\in \mathbb{N}$. Assuming $p=0$, this leads to the condition :
$$ n_{z0} = - \frac{\Delta\Phi}{k_0 \Delta z} $$
Let's define the following function:
```
def ideal_spectrum(b,e,phi,N=6,f=3.7e9):
nz = np.arange(-10,10,0.1)
k0 = (2*pi*f)/c
PHI = k0*nz*(b+e) + phi
dP = sinc(k0*nz*b/2)*(sin(N*PHI/2)/sin(PHI/2))**2
return(nz,dP)
```
And trace the spectrum of an ideal launcher:
```
nz, dP_ideal = ideal_spectrum(b=8e-3, e=2e-3, phi=pi/2, N=6)
plot(nz, abs(dP_ideal), lw=2)
xlabel('$n_z=k_z/k_0$');
ylabel('Power density [a.u.]');
title('Power density spectrum of an ideal LH grill launcher')
grid('on')
plt.savefig('multijunction_ideal_spectrum.png', dpi=150)
```
## Realistic grill
Let's illustrate this with a more realistic case. Below we define a function that generate the electric field along z for N waveguides, d-spaced and with a constant phase shift. The spacial precision can be set optionnaly.
```
def generate_Efield(b,e,phi,N=6,dz=1e-4,A=1):
# generate the z-axis, between [z_min, z_max[ by dz steps
z_min = 0 - 0.01
z_max = N*(b+e) + 0.01
z = arange(z_min, z_max, dz)
# construct the Efield (complex valued)
E = zeros_like(z,dtype=complex)
for idx in arange(N):
E[ (z>=idx*(b+e)) & (z < idx*(b+e)+b) ] = A * exp(1j*idx*phi)
return(z,E)
```
Then we use this function to generate the electric field at the mouth of a 6 waveguide launcher, of waveguide width b=8mm, septum thickness e=2mm with a phase shift between waveguides of 90° ($\pi/2$) :
```
z,E = generate_Efield(b=8e-3, e=2e-3,phi=pi/2)
fig, (ax1, ax2) = plt.subplots(2,1,sharex=True)
ax1.plot(z,abs(E), lw=2)
ax1.set_ylabel('Amplitude [a.u.]')
ax1.grid(True)
ax1.set_ylim((-0.1,1.1))
ax2.plot(z,angle(E)*180/pi,'g', lw=2)
ax2.set_xlabel('z [m]')
ax2.set_ylabel('Phase [deg]')
ax2.grid(True)
ax2.set_ylim((-200, 200))
fig.savefig('multijunction_ideal_excitation.png', dpi=150)
```
Now, let's take the fourier transform of such a field (the source frequency is here f=3.7 GHz, the frequency of the Tore Supra LH system).
```
def calculate_spectrum(z,E,f=3.7e9):
k0 = 2*pi*f/c
lambda0 = c/f
# fourier domain points
B = 2**18
Efft = np.fft.fftshift(np.fft.fft(E,B))
# fourier domain bins
dz = z[1] - z[0] # assumes spatial period is constant
df = 1/(B*dz)
K = arange(-B/2,+B/2)
# spatial frequency bins
Fz= K*df
# parallel index is kz/k0
nz= (2*pi/k0)*Fz
# ~ power density spectrum
p = (dz)**2/lambda0 * (1/2*Efft*conj(Efft));
return(nz,p)
nz,p = calculate_spectrum(z,E)
plot(nz,real(p),lw=2)
xlim((-10,10))
xlabel('$n_z=k_z/k_0$')
ylabel('Power density [a.u.]')
title('Spectral power density of an ideal LH launcher')
grid('on')
plt.savefig('multijunction_ideal_spectrum.png', dpi=150)
```
The main component of the spectrum seems located near $n_z$=2. Looking at the previous analytical formula, which expresses the extremum of the power density spectrum:
```
f = 3.7e9 # frequency [Hz]
b = 8e-3 # waveguide width [m]
e = 2e-3 # septum width [m]
k0 = (2*pi*f) / c # wavenumber in vacuum
delta_phi = pi/2 # phase shift between waveguides
```
$$n_{z0} = \frac{k_{z0}}{k_0} = \frac{\Delta \Phi}{k_0 \Delta z }$$
```
nz0 = pi/2 / ((b+e) * k0) # main component of the spectrum
nz0
```
Which is what we expected from the previous Figure.
## Current Drive Direction
Let's assume that the plasma current is clockwise as seen from the top of a tokamak. Let's also define the positive direction of the toroidal magnetic field being in the same direction than the plasma current, i.e. :
$$
\mathbf{B}_0 = B_0 \mathbf{e}_\parallel
$$
and
$$
\mathbf{I}_p = I_p \mathbf{e}_\parallel
$$
Let be $\mathbf{J}_{LH}$ the current density created by the Lower Hybrid system. The current density is expressed as
$$
\mathbf{J}_{LH} = - n e v_\parallel \mathbf{e}_\parallel
$$
where $v_\parallel=c/n_\parallel$. As we want the current driven to be in the same direction than the plasma current, i.e.:
$$
\mathbf{J}_{LH}\cdot\mathbf{I}_p > 0
$$
one must have :
$$
n_\parallel < 0
$$
## RF Measurements of the multijunction
```
import numpy as np
calibration = np.loadtxt('LH2_Multijunction_data/calibration.s2p', skiprows=5)
fwd1 = np.loadtxt('LH2_Multijunction_data/fwd1.s2p', skiprows=5)
fwd2 = np.loadtxt('LH2_Multijunction_data/fwd2.s2p', skiprows=5)
fwd3 = np.loadtxt('LH2_Multijunction_data/fwd3.s2p', skiprows=5)
fwd4 = np.loadtxt('LH2_Multijunction_data/fwd4.s2p', skiprows=5)
fwd5 = np.loadtxt('LH2_Multijunction_data/fwd5.s2p', skiprows=5)
fwd6 = np.loadtxt('LH2_Multijunction_data/fwd6.s2p', skiprows=5)
f = calibration[:,0]
S21_cal = calibration[:,3]
fig, ax = plt.subplots()
ax.plot(f/1e9, fwd1[:,3]-S21_cal)
ax.plot(f/1e9, fwd2[:,3]-S21_cal)
ax.plot(f/1e9, fwd3[:,3]-S21_cal)
ax.plot(f/1e9, fwd4[:,3]-S21_cal)
ax.plot(f/1e9, fwd5[:,3]-S21_cal)
#ax.plot(f/1e9, fwd6[:,3]-S21_cal)
ax.legend(('Forward wg#1', 'Forward wg#2','Forward wg#3','Forward wg#4','Forward wg#5'))
ax.grid(True)
ax.axvline(3.7, color='r', ls='--')
ax.axhline(10*np.log10(1/6), color='gray', ls='--')
ax.set_ylabel('S21 [dB]')
ax.set_xlabel('f [GHz]')
fwds = [fwd1, fwd2, fwd3, fwd4, fwd5, fwd6]
s21 = []
for fwd in fwds:
s21.append(10**((fwd[:,3]-S21_cal)/20) * np.exp(1j*fwd[:,4]*np.pi/180))
s21 = np.asarray(s21)
# find the 3.7 GHz point
idx = np.argwhere(f == 3.7e9)
s21_3dot7 = s21[:,idx].squeeze()
fig, ax = plt.subplots(nrows=2)
ax[0].bar(np.arange(1,7), np.abs(s21_3dot7))
ax[1].bar(np.arange(1,7), 180/np.pi*np.unwrap(np.angle(s21_3dot7)))
```
Clearly, the last waveguide measurements are strange...
## Checking the power conservation
```
fig, ax = plt.subplots()
plot(f/1e9, abs(s21**2).sum(axis=0))
ax.grid(True)
ax.axvline(3.7, color='r', ls='--')
ax.set_ylabel('$\sum |S_{21}|^2$ ')
ax.set_xlabel('f [GHz]')
```
Clearly, the power conservation is far from being verified, because of the large incertitude of the measurement here.
### CSS Styling
```
from IPython.core.display import HTML
def css_styling():
styles = open("../../styles/custom.css", "r").read()
return HTML(styles)
css_styling()
```
|
github_jupyter
|
%pylab
%matplotlib inline
from scipy.constants import c
def ideal_spectrum(b,e,phi,N=6,f=3.7e9):
nz = np.arange(-10,10,0.1)
k0 = (2*pi*f)/c
PHI = k0*nz*(b+e) + phi
dP = sinc(k0*nz*b/2)*(sin(N*PHI/2)/sin(PHI/2))**2
return(nz,dP)
nz, dP_ideal = ideal_spectrum(b=8e-3, e=2e-3, phi=pi/2, N=6)
plot(nz, abs(dP_ideal), lw=2)
xlabel('$n_z=k_z/k_0$');
ylabel('Power density [a.u.]');
title('Power density spectrum of an ideal LH grill launcher')
grid('on')
plt.savefig('multijunction_ideal_spectrum.png', dpi=150)
def generate_Efield(b,e,phi,N=6,dz=1e-4,A=1):
# generate the z-axis, between [z_min, z_max[ by dz steps
z_min = 0 - 0.01
z_max = N*(b+e) + 0.01
z = arange(z_min, z_max, dz)
# construct the Efield (complex valued)
E = zeros_like(z,dtype=complex)
for idx in arange(N):
E[ (z>=idx*(b+e)) & (z < idx*(b+e)+b) ] = A * exp(1j*idx*phi)
return(z,E)
z,E = generate_Efield(b=8e-3, e=2e-3,phi=pi/2)
fig, (ax1, ax2) = plt.subplots(2,1,sharex=True)
ax1.plot(z,abs(E), lw=2)
ax1.set_ylabel('Amplitude [a.u.]')
ax1.grid(True)
ax1.set_ylim((-0.1,1.1))
ax2.plot(z,angle(E)*180/pi,'g', lw=2)
ax2.set_xlabel('z [m]')
ax2.set_ylabel('Phase [deg]')
ax2.grid(True)
ax2.set_ylim((-200, 200))
fig.savefig('multijunction_ideal_excitation.png', dpi=150)
def calculate_spectrum(z,E,f=3.7e9):
k0 = 2*pi*f/c
lambda0 = c/f
# fourier domain points
B = 2**18
Efft = np.fft.fftshift(np.fft.fft(E,B))
# fourier domain bins
dz = z[1] - z[0] # assumes spatial period is constant
df = 1/(B*dz)
K = arange(-B/2,+B/2)
# spatial frequency bins
Fz= K*df
# parallel index is kz/k0
nz= (2*pi/k0)*Fz
# ~ power density spectrum
p = (dz)**2/lambda0 * (1/2*Efft*conj(Efft));
return(nz,p)
nz,p = calculate_spectrum(z,E)
plot(nz,real(p),lw=2)
xlim((-10,10))
xlabel('$n_z=k_z/k_0$')
ylabel('Power density [a.u.]')
title('Spectral power density of an ideal LH launcher')
grid('on')
plt.savefig('multijunction_ideal_spectrum.png', dpi=150)
f = 3.7e9 # frequency [Hz]
b = 8e-3 # waveguide width [m]
e = 2e-3 # septum width [m]
k0 = (2*pi*f) / c # wavenumber in vacuum
delta_phi = pi/2 # phase shift between waveguides
nz0 = pi/2 / ((b+e) * k0) # main component of the spectrum
nz0
import numpy as np
calibration = np.loadtxt('LH2_Multijunction_data/calibration.s2p', skiprows=5)
fwd1 = np.loadtxt('LH2_Multijunction_data/fwd1.s2p', skiprows=5)
fwd2 = np.loadtxt('LH2_Multijunction_data/fwd2.s2p', skiprows=5)
fwd3 = np.loadtxt('LH2_Multijunction_data/fwd3.s2p', skiprows=5)
fwd4 = np.loadtxt('LH2_Multijunction_data/fwd4.s2p', skiprows=5)
fwd5 = np.loadtxt('LH2_Multijunction_data/fwd5.s2p', skiprows=5)
fwd6 = np.loadtxt('LH2_Multijunction_data/fwd6.s2p', skiprows=5)
f = calibration[:,0]
S21_cal = calibration[:,3]
fig, ax = plt.subplots()
ax.plot(f/1e9, fwd1[:,3]-S21_cal)
ax.plot(f/1e9, fwd2[:,3]-S21_cal)
ax.plot(f/1e9, fwd3[:,3]-S21_cal)
ax.plot(f/1e9, fwd4[:,3]-S21_cal)
ax.plot(f/1e9, fwd5[:,3]-S21_cal)
#ax.plot(f/1e9, fwd6[:,3]-S21_cal)
ax.legend(('Forward wg#1', 'Forward wg#2','Forward wg#3','Forward wg#4','Forward wg#5'))
ax.grid(True)
ax.axvline(3.7, color='r', ls='--')
ax.axhline(10*np.log10(1/6), color='gray', ls='--')
ax.set_ylabel('S21 [dB]')
ax.set_xlabel('f [GHz]')
fwds = [fwd1, fwd2, fwd3, fwd4, fwd5, fwd6]
s21 = []
for fwd in fwds:
s21.append(10**((fwd[:,3]-S21_cal)/20) * np.exp(1j*fwd[:,4]*np.pi/180))
s21 = np.asarray(s21)
# find the 3.7 GHz point
idx = np.argwhere(f == 3.7e9)
s21_3dot7 = s21[:,idx].squeeze()
fig, ax = plt.subplots(nrows=2)
ax[0].bar(np.arange(1,7), np.abs(s21_3dot7))
ax[1].bar(np.arange(1,7), 180/np.pi*np.unwrap(np.angle(s21_3dot7)))
fig, ax = plt.subplots()
plot(f/1e9, abs(s21**2).sum(axis=0))
ax.grid(True)
ax.axvline(3.7, color='r', ls='--')
ax.set_ylabel('$\sum |S_{21}|^2$ ')
ax.set_xlabel('f [GHz]')
from IPython.core.display import HTML
def css_styling():
styles = open("../../styles/custom.css", "r").read()
return HTML(styles)
css_styling()
| 0.523908 | 0.993183 |
# Tutorial Python
## Print
```
print("Halo, nama saya Adam, biasa dipanggil Arthur")
print('Halo, nama saya Adam, biasa dipanggil Arthur')
print("""Halo, nama saya Adam, biasa dipanggil Arthur""")
print('''Halo, nama saya Adam, biasa dipanggil Arthur''')
```
## Variable
```
a = 2
a
a, b = 3, 4
a
b
a, b, c = 6, 8, 10
a
b
c
a, b, c
```
## List
```
a = [1, 2, 3, 'empat', 'lima', True]
a[0]
a[2]
a[-2]
a[-1]
a[-6]
a.append(7)
a
len(a)
```
## Slicing
`inclusive` : `exclusive` : `step`
```
a = [0, 1, 2, 3, 4, 5, 6, 7, 8]
a[0:2]
a[0:5]
a[0:-3]
a[:]
a[:5]
a[::]
a[1:8:2]
a[::2]
a[::-2]
```
## Join & Split
```
a = ['cat', 'dog', 'fish']
" ".join(a)
", ".join(a)
a = "Halo nama saya Adam biasa dipanggil Arthur"
a.split()
a = "Halo, nama saya Adam, biasa dipanggil Arthur"
a.split(", ")
```
## Dictionary
```
a = {
'cat': 'kucing',
'dog': 'anjing',
'fish': 'ikan',
}
a
a['cat']
a['bird'] = 'burung'
a['elephant'] = 'gajah'
a
a.keys()
a.values()
nilai = {'naruto': 40, 'kaneki': 80, 'izuku': 85, 'ayanokouji': 100}
nilai
nilai['ayanokouji'] = 90
nilai
```
## Function
```
def jumlah(a, b):
return a + b
jumlah(5,6)
def kali(a, b):
return a * b
kali(5,6)
def pangkat(a, b):
return a ** b
pangkat(5, 6)
```
## Conditionals
```
score = 50
def konversi_indeks(indeks):
if 0 <= indeks < 60:
return "D"
elif 60 <= indeks < 70:
return "C"
elif 70 <= indeks < 80:
return "B"
elif 80 <= indeks < 90:
return "A"
elif 90 <= indeks <= 100:
return "S"
else:
return "Nilai nya ngaco"
konversi_indeks(60)
konversi_indeks(100)
konversi_indeks(1234)
```
## Iterasi
```
numbers = [1, 2, 3, 4, 5, 6, 7]
for n in numbers:
print(n)
for score in [50, 60, 70, 80, 90, 100, 110]:
print("Indeks: ", konversi_indeks(score))
animals = ['Cat', 'Fish', 'Dog', 'Elephant', 'Bird']
for animal in animals:
print(animal)
for animal in animals:
print(animal.upper())
for animal in animals:
print(animal.lower())
for angka in range(2, 10, 2):
print(angka)
animals ={
'Cat': 'Kucing',
'Fish': 'Ikan',
'Elephant': 'Gajah',
'Dog': 'Anjing',
'Bird': 'Burung'
}
for animal in animals.keys():
print(animal)
for animal in animals.values():
print(animal)
for animal in animals.keys():
print(f"Bahasa Indonesia dari {animal} adalah {animals[animal]}")
a = []
for angka in range(8):
a.append(angka ** 2)
a
```
## Python Comprehension
```
b = [angka ** 2 for angka in range(10)]
b
b = {angka: angka ** 2 for angka in range(10)}
b
{1, 1, 2, 2, 3, 3, 4, 4, 5, 5}
```
|
github_jupyter
|
print("Halo, nama saya Adam, biasa dipanggil Arthur")
print('Halo, nama saya Adam, biasa dipanggil Arthur')
print("""Halo, nama saya Adam, biasa dipanggil Arthur""")
print('''Halo, nama saya Adam, biasa dipanggil Arthur''')
a = 2
a
a, b = 3, 4
a
b
a, b, c = 6, 8, 10
a
b
c
a, b, c
a = [1, 2, 3, 'empat', 'lima', True]
a[0]
a[2]
a[-2]
a[-1]
a[-6]
a.append(7)
a
len(a)
a = [0, 1, 2, 3, 4, 5, 6, 7, 8]
a[0:2]
a[0:5]
a[0:-3]
a[:]
a[:5]
a[::]
a[1:8:2]
a[::2]
a[::-2]
a = ['cat', 'dog', 'fish']
" ".join(a)
", ".join(a)
a = "Halo nama saya Adam biasa dipanggil Arthur"
a.split()
a = "Halo, nama saya Adam, biasa dipanggil Arthur"
a.split(", ")
a = {
'cat': 'kucing',
'dog': 'anjing',
'fish': 'ikan',
}
a
a['cat']
a['bird'] = 'burung'
a['elephant'] = 'gajah'
a
a.keys()
a.values()
nilai = {'naruto': 40, 'kaneki': 80, 'izuku': 85, 'ayanokouji': 100}
nilai
nilai['ayanokouji'] = 90
nilai
def jumlah(a, b):
return a + b
jumlah(5,6)
def kali(a, b):
return a * b
kali(5,6)
def pangkat(a, b):
return a ** b
pangkat(5, 6)
score = 50
def konversi_indeks(indeks):
if 0 <= indeks < 60:
return "D"
elif 60 <= indeks < 70:
return "C"
elif 70 <= indeks < 80:
return "B"
elif 80 <= indeks < 90:
return "A"
elif 90 <= indeks <= 100:
return "S"
else:
return "Nilai nya ngaco"
konversi_indeks(60)
konversi_indeks(100)
konversi_indeks(1234)
numbers = [1, 2, 3, 4, 5, 6, 7]
for n in numbers:
print(n)
for score in [50, 60, 70, 80, 90, 100, 110]:
print("Indeks: ", konversi_indeks(score))
animals = ['Cat', 'Fish', 'Dog', 'Elephant', 'Bird']
for animal in animals:
print(animal)
for animal in animals:
print(animal.upper())
for animal in animals:
print(animal.lower())
for angka in range(2, 10, 2):
print(angka)
animals ={
'Cat': 'Kucing',
'Fish': 'Ikan',
'Elephant': 'Gajah',
'Dog': 'Anjing',
'Bird': 'Burung'
}
for animal in animals.keys():
print(animal)
for animal in animals.values():
print(animal)
for animal in animals.keys():
print(f"Bahasa Indonesia dari {animal} adalah {animals[animal]}")
a = []
for angka in range(8):
a.append(angka ** 2)
a
b = [angka ** 2 for angka in range(10)]
b
b = {angka: angka ** 2 for angka in range(10)}
b
{1, 1, 2, 2, 3, 3, 4, 4, 5, 5}
| 0.299617 | 0.930962 |
```
import Brunel
import datetime
import random
import pandas as pd
nodes = pd.read_csv("input/nodes.csv")
edges = pd.read_csv("input/edges.csv")
nodes
edges
```
Adding some random dates so that I can test the temporal controls
```
days = datetime.timedelta(days=1)
weeks = datetime.timedelta(weeks=1)
months = 30*days
years = datetime.timedelta(days=365)
def random_date(start=datetime.date(year=1800, month=1, day=1), end=datetime.date(year=1890, month=12, day=31),
within=None):
if within:
start = within[0]
end = within[1]
start = datetime.datetime.combine(start, datetime.time()).timestamp()
end = datetime.datetime.combine(end, datetime.time()).timestamp()
result = start + random.random() * (end - start)
return datetime.datetime.fromtimestamp(result).date()
def random_duration(start=datetime.date(year=1800, month=1, day=1),
end=datetime.date(year=1890, month=12, day=31),
within=None,
minimum=20*years, maximum=80*years,
breach_maximum=False):
if within:
start = within[0]
end = within[1]
mindur = minimum.total_seconds()
maxdur = maximum.total_seconds()
start = random_date(start=start, end=end)
if not breach_maximum:
lifedur = (end - start).total_seconds()
if maxdur > lifedur:
maxdur = lifedur
if mindur > maxdur:
mindur = 0.5*maxdur
dur = mindur + random.random() * (maxdur-mindur)
return (start, start + datetime.timedelta(seconds=dur))
def random_lifetime(start=datetime.date(year=1800, month=1, day=1),
end=datetime.date(year=1890, month=12, day=31),
maximum_age=80*years, all_adults=True):
if all_adults:
minimum = 18*years
else:
minimum = 1*day
return random_duration(start=start, end=end, minimum=minimum, maximum=maximum_age, breach_maximum=True)
def adult(lifetime):
start = lifetime[0]
end = lifetime[1]
start = start + 18*years
if start > end:
raise ValueError("Not an adult %s => %s" % (start.isoformat(), end.isoformat()))
return (start, end)
lifetime = random_lifetime()
print(lifetime)
print(adult(lifetime))
print(random_duration(within=adult(lifetime), minimum=6*months, maximum=5*years))
print(f"Lived {(lifetime[1]-lifetime[0]).total_seconds() / (3600*24*365)} years")
def get_earliest(start, end, ids):
try:
start = ids[start][0]
except KeyError:
try:
return ids[end][0]
except KeyError:
return None
try:
end = ids[end][0]
except KeyError:
return start
if start < end:
return end
else:
return start
def get_latest(start, end, ids):
try:
start = ids[start][1]
except KeyError:
try:
return ids[end][1]
except KeyError:
return None
try:
end = ids[end][1]
except KeyError:
return start
if start < end:
return start
else:
return end
Brunel.DateRange(start=lifetime[0], end=lifetime[1])
lifetimes = {}
def add_random_dates_to_node(node):
lifetime = random_lifetime()
duration = lifetime
if "alive" in node.state:
node.state["alive"] = Brunel.DateRange(start=lifetime[0], end=lifetime[1])
lifetimes[node.getID()] = lifetime
duration = adult(lifetime)
if "positions" in node.state:
pos = node.state["positions"]
for key in pos.keys():
member = random_duration(within=duration, minimum=6*months, maximum=20*years)
pos[key] = Brunel.DateRange(start=member[0], end=member[1])
node.state["positions"] = pos
if "affiliations" in node.state:
aff = node.state["affiliations"]
for key in aff.keys():
member = random_duration(within=duration, minimum=5*years, maximum=20*years)
aff[key] = Brunel.DateRange(start=member[0], end=member[1])
node.state["affiliations"] = aff
return node
def add_random_dates_to_message(message):
start = get_earliest(message.getSender(), message.getReceiver(), lifetimes)
end = get_latest(message.getSender(), message.getReceiver(), lifetimes)
if not start:
start = datetime.date(year=1850, month=1, day=1)
end = datetime.date(year=1870, month=12, day=31)
if not end:
end = start + 12*months
sent = random_date(start=start, end=end)
message.state["sent"] = Brunel.DateRange(start=sent, end=sent)
return message
social = Brunel.Social.load_from_csv("input/nodes.csv", "input/edges.csv",
modifiers={"person": add_random_dates_to_node,
"business": add_random_dates_to_node,
"message": add_random_dates_to_message})
with open("data.json", "w") as FILE:
FILE.write(Brunel.stringify(social))
```
|
github_jupyter
|
import Brunel
import datetime
import random
import pandas as pd
nodes = pd.read_csv("input/nodes.csv")
edges = pd.read_csv("input/edges.csv")
nodes
edges
days = datetime.timedelta(days=1)
weeks = datetime.timedelta(weeks=1)
months = 30*days
years = datetime.timedelta(days=365)
def random_date(start=datetime.date(year=1800, month=1, day=1), end=datetime.date(year=1890, month=12, day=31),
within=None):
if within:
start = within[0]
end = within[1]
start = datetime.datetime.combine(start, datetime.time()).timestamp()
end = datetime.datetime.combine(end, datetime.time()).timestamp()
result = start + random.random() * (end - start)
return datetime.datetime.fromtimestamp(result).date()
def random_duration(start=datetime.date(year=1800, month=1, day=1),
end=datetime.date(year=1890, month=12, day=31),
within=None,
minimum=20*years, maximum=80*years,
breach_maximum=False):
if within:
start = within[0]
end = within[1]
mindur = minimum.total_seconds()
maxdur = maximum.total_seconds()
start = random_date(start=start, end=end)
if not breach_maximum:
lifedur = (end - start).total_seconds()
if maxdur > lifedur:
maxdur = lifedur
if mindur > maxdur:
mindur = 0.5*maxdur
dur = mindur + random.random() * (maxdur-mindur)
return (start, start + datetime.timedelta(seconds=dur))
def random_lifetime(start=datetime.date(year=1800, month=1, day=1),
end=datetime.date(year=1890, month=12, day=31),
maximum_age=80*years, all_adults=True):
if all_adults:
minimum = 18*years
else:
minimum = 1*day
return random_duration(start=start, end=end, minimum=minimum, maximum=maximum_age, breach_maximum=True)
def adult(lifetime):
start = lifetime[0]
end = lifetime[1]
start = start + 18*years
if start > end:
raise ValueError("Not an adult %s => %s" % (start.isoformat(), end.isoformat()))
return (start, end)
lifetime = random_lifetime()
print(lifetime)
print(adult(lifetime))
print(random_duration(within=adult(lifetime), minimum=6*months, maximum=5*years))
print(f"Lived {(lifetime[1]-lifetime[0]).total_seconds() / (3600*24*365)} years")
def get_earliest(start, end, ids):
try:
start = ids[start][0]
except KeyError:
try:
return ids[end][0]
except KeyError:
return None
try:
end = ids[end][0]
except KeyError:
return start
if start < end:
return end
else:
return start
def get_latest(start, end, ids):
try:
start = ids[start][1]
except KeyError:
try:
return ids[end][1]
except KeyError:
return None
try:
end = ids[end][1]
except KeyError:
return start
if start < end:
return start
else:
return end
Brunel.DateRange(start=lifetime[0], end=lifetime[1])
lifetimes = {}
def add_random_dates_to_node(node):
lifetime = random_lifetime()
duration = lifetime
if "alive" in node.state:
node.state["alive"] = Brunel.DateRange(start=lifetime[0], end=lifetime[1])
lifetimes[node.getID()] = lifetime
duration = adult(lifetime)
if "positions" in node.state:
pos = node.state["positions"]
for key in pos.keys():
member = random_duration(within=duration, minimum=6*months, maximum=20*years)
pos[key] = Brunel.DateRange(start=member[0], end=member[1])
node.state["positions"] = pos
if "affiliations" in node.state:
aff = node.state["affiliations"]
for key in aff.keys():
member = random_duration(within=duration, minimum=5*years, maximum=20*years)
aff[key] = Brunel.DateRange(start=member[0], end=member[1])
node.state["affiliations"] = aff
return node
def add_random_dates_to_message(message):
start = get_earliest(message.getSender(), message.getReceiver(), lifetimes)
end = get_latest(message.getSender(), message.getReceiver(), lifetimes)
if not start:
start = datetime.date(year=1850, month=1, day=1)
end = datetime.date(year=1870, month=12, day=31)
if not end:
end = start + 12*months
sent = random_date(start=start, end=end)
message.state["sent"] = Brunel.DateRange(start=sent, end=sent)
return message
social = Brunel.Social.load_from_csv("input/nodes.csv", "input/edges.csv",
modifiers={"person": add_random_dates_to_node,
"business": add_random_dates_to_node,
"message": add_random_dates_to_message})
with open("data.json", "w") as FILE:
FILE.write(Brunel.stringify(social))
| 0.382487 | 0.42471 |
# Probability concepts using Python
### Dr. Tirthajyoti Sarkar, Fremont, CA 94536
---
This notebook illustrates the concept of probability (frequntist definition) using simple scripts and functions.
## Set theory basics
Set theory is a branch of mathematical logic that studies sets, which informally are collections of objects. Although any type of object can be collected into a set, set theory is applied most often to objects that are relevant to mathematics. The language of set theory can be used in the definitions of nearly all mathematical objects.
**Set theory is commonly employed as a foundational system for modern mathematics**.
Python offers a **native data structure called set**, which can be used as a proxy for a mathematical set for almost all purposes.
```
# Directly with curly braces
Set1 = {1,2}
print (Set1)
print(type(Set1))
my_list=[1,2,3,4]
my_set_from_list = set(my_list)
print(my_set_from_list)
```
### Membership testing with `in` and `not in`
```
my_set = set([1,3,5])
print("Here is my set:",my_set)
print("1 is in the set:",1 in my_set)
print("2 is in the set:",2 in my_set)
print("4 is NOT in the set:",4 not in my_set)
```
### Set relations
* **Subset**
* **Superset**
* **Disjoint**
* **Universal set**
* **Null set**
```
Univ = set([x for x in range(11)])
Super = set([x for x in range(11) if x%2==0])
disj = set([x for x in range(11) if x%2==1])
Sub = set([4,6])
Null = set([x for x in range(11) if x>10])
print("Universal set (all the positive integers up to 10):",Univ)
print("All the even positive integers up to 10:",Super)
print("All the odd positive integers up to 10:",disj)
print("Set of 2 elements, 4 and 6:",Sub)
print("A null set:", Null)
print('Is "Super" a superset of "Sub"?',Super.issuperset(Sub))
print('Is "Super" a subset of "Univ"?',Super.issubset(Univ))
print('Is "Sub" a superset of "Super"?',Sub.issuperset(Super))
print('Is "Super" disjoint with "disj"?',Sub.isdisjoint(disj))
```
### Set algebra/Operations
* **Equality**
* **Intersection**
* **Union**
* **Complement**
* **Difference**
* **Cartesian product**
```
S1 = {1,2}
S2 = {2,2,1,1,2}
print ("S1 and S2 are equal because order or repetition of elements do not matter for sets\nS1==S2:", S1==S2)
S1 = {1,2,3,4,5,6}
S2 = {1,2,3,4,0,6}
print ("S1 and S2 are NOT equal because at least one element is different\nS1==S2:", S1==S2)
```
In mathematics, the intersection A ∩ B of two sets A and B is the set that contains all elements of A that also belong to B (or equivalently, all elements of B that also belong to A), but no other elements. Formally,
$$ {\displaystyle A\cap B=\{x:x\in A{\text{ and }}x\in B\}.} $$

```
# Define a set using list comprehension
S1 = set([x for x in range(1,11) if x%3==0])
print("S1:", S1)
S2 = set([x for x in range(1,7)])
print("S2:", S2)
# Both intersection method or & can be used
S_intersection = S1.intersection(S2)
print("Intersection of S1 and S2:", S_intersection)
S_intersection = S1 & S2
print("Intersection of S1 and S2:", S_intersection)
S3 = set([x for x in range(6,10)])
print("S3:", S3)
S1_S2_S3 = S1.intersection(S2).intersection(S3)
print("Intersection of S1, S2, and S3:", S1_S2_S3)
```
In set theory, the union (denoted by ∪) of a collection of sets is the set of all elements in the collection. It is one of the fundamental operations through which sets can be combined and related to each other. Formally,
$$ {A\cup B=\{x:x\in A{\text{ or }}x\in B\}} $$

```
# Both union method or | can be used
S1 = set([x for x in range(1,11) if x%3==0])
print("S1:", S1)
S2 = set([x for x in range(1,5)])
print("S2:", S2)
S_union = S1.union(S2)
print("Union of S1 and S2:", S_union)
S_union = S1 | S2
print("Union of S1 and S2:", S_union)
```
### Set algebra laws
**Commutative law:**
$$ {\displaystyle A\cap B=B\cap A} $$
$$ {\displaystyle A\cup (B\cup C)=(A\cup B)\cup C} $$
**Associative law:**
$$ {\displaystyle (A\cap B)\cap C=A\cap (B\cap C)} $$
$$ {\displaystyle A\cap (B\cup C)=(A\cap B)\cup (A\cap C)} $$
**Distributive law:**
$$ {\displaystyle A\cap (B\cup C)=(A\cap B)\cup (A\cap C)} $$
$$ {\displaystyle A\cup (B\cap C)=(A\cup B)\cap (A\cup C)} $$
### Complement
If A is a set, then the absolute complement of A (or simply the complement of A) is the set of elements not in A. In other words, if U is the universe that contains all the elements under study, and there is no need to mention it because it is obvious and unique, then the absolute complement of A is the relative complement of A in U. Formally,
$$ {\displaystyle A^{\complement }=\{x\in U\mid x\notin A\}.} $$
You can take the union of two sets and if that is equal to the universal set (in the context of your problem), then you have found the right complement
```
S=set([x for x in range (21) if x%2==0])
print ("S is the set of even numbers between 0 and 20:", S)
S_complement = set([x for x in range (21) if x%2!=0])
print ("S_complement is the set of odd numbers between 0 and 20:", S_complement)
print ("Is the union of S and S_complement equal to all numbers between 0 and 20?",
S.union(S_complement)==set([x for x in range (21)]))
```
**De Morgan's laws**
$$ {\displaystyle \left(A\cup B\right)^{\complement }=A^{\complement }\cap B^{\complement }.} $$
$$ {\displaystyle \left(A\cap B\right)^{\complement }=A^{\complement }\cup B^{\complement }.} $$
**Complement laws**
$$ {\displaystyle A\cup A^{\complement }=U.} $$
$$ {\displaystyle A\cap A^{\complement }=\varnothing .} $$
$$ {\displaystyle \varnothing ^{\complement }=U.} $$
$$ {\displaystyle U^{\complement }=\varnothing .} $$
$$ {\displaystyle {\text{If }}A\subset B{\text{, then }}B^{\complement }\subset A^{\complement }.} $$
### Difference between sets
If A and B are sets, then the relative complement of A in B, also termed the set-theoretic difference of B and A, is the **set of elements in B but not in A**.
$$ {\displaystyle B\setminus A=\{x\in B\mid x\notin A\}.} $$

```
S1 = set([x for x in range(31) if x%3==0])
print ("Set S1:", S1)
S2 = set([x for x in range(31) if x%5==0])
print ("Set S2:", S2)
S_difference = S2-S1
print("Difference of S1 and S2 i.e. S2\S1:", S_difference)
S_difference = S1.difference(S2)
print("Difference of S2 and S1 i.e. S1\S2:", S_difference)
```
**Following identities can be obtained with algebraic manipulation: **
$$ {\displaystyle C\setminus (A\cap B)=(C\setminus A)\cup (C\setminus B)} $$
$$ {\displaystyle C\setminus (A\cup B)=(C\setminus A)\cap (C\setminus B)} $$
$$ {\displaystyle C\setminus (B\setminus A)=(C\cap A)\cup (C\setminus B)} $$
$$ {\displaystyle C\setminus (C\setminus A)=(C\cap A)} $$
$$ {\displaystyle (B\setminus A)\cap C=(B\cap C)\setminus A=B\cap (C\setminus A)} $$
$$ {\displaystyle (B\setminus A)\cup C=(B\cup C)\setminus (A\setminus C)} $$
$$ {\displaystyle A\setminus A=\emptyset} $$
$$ {\displaystyle \emptyset \setminus A=\emptyset } $$
$$ {\displaystyle A\setminus \emptyset =A} $$
$$ {\displaystyle A\setminus U=\emptyset } $$
### Symmetric difference
In set theory, the ***symmetric difference***, also known as the ***disjunctive union***, of two sets is the set of elements which are in either of the sets and not in their intersection.
$$ {\displaystyle A\,\triangle \,B=\{x:(x\in A)\oplus (x\in B)\}}$$
$$ {\displaystyle A\,\triangle \,B=(A\smallsetminus B)\cup (B\smallsetminus A)} $$
$${\displaystyle A\,\triangle \,B=(A\cup B)\smallsetminus (A\cap B)} $$
**Some properties,**
$$ {\displaystyle A\,\triangle \,B=B\,\triangle \,A,} $$
$$ {\displaystyle (A\,\triangle \,B)\,\triangle \,C=A\,\triangle \,(B\,\triangle \,C).} $$
**The empty set is neutral, and every set is its own inverse:**
$$ {\displaystyle A\,\triangle \,\varnothing =A,} $$
$$ {\displaystyle A\,\triangle \,A=\varnothing .} $$
```
print("S1",S1)
print("S2",S2)
print("Symmetric difference", S1^S2)
print("Symmetric difference", S2.symmetric_difference(S1))
```
### Cartesian product
In set theory (and, usually, in other parts of mathematics), a Cartesian product is a mathematical operation that returns a set (or product set or simply product) from multiple sets. That is, for sets A and B, the Cartesian product A × B is the set of all ordered pairs (a, b) where a ∈ A and b ∈ B.
$$ {\displaystyle A\times B=\{\,(a,b)\mid a\in A\ {\mbox{ and }}\ b\in B\,\}.} $$
More generally, a Cartesian product of n sets, also known as an n-fold Cartesian product, can be represented by an array of n dimensions, where each element is an *n-tuple*. An ordered pair is a *2-tuple* or couple. The Cartesian product is named after [René Descartes](https://en.wikipedia.org/wiki/Ren%C3%A9_Descartes) whose formulation of analytic geometry gave rise to the concept.
```
A = set(['a','b','c'])
S = {1,2,3}
def cartesian_product(S1,S2):
result = set()
for i in S1:
for j in S2:
result.add(tuple([i,j]))
return (result)
C = cartesian_product(A,S)
print("Cartesian product of A and S\n{} X {}:{}".format(A,S,C))
print("Length of the Cartesian product set:",len(C))
```
Note that because these are ordered pairs, **same element can be repeated inside the pair** i.e. even if two sets contain some identical elements, they can be paired up in the Cartesian product.
Instead of writing functions ourselves, we could use the **`itertools`** library of Python. Remember to **turn the resulting product object** into a list for viewing and subsequent processing.
```
from itertools import product as prod
A = set([x for x in range(1,7)])
B = set([x for x in range(1,7)])
p=list(prod(A,B))
print("A is set of all possible throws of a dice:",A)
print("B is set of all possible throws of a dice:",B)
print ("\nProduct of A and B is the all possible combinations of A and B thrown together:\n",p)
```
### Cartesian Power
The Cartesian square (or binary Cartesian product) of a set X is the Cartesian product $X^2 = X × X$. An example is the 2-dimensional plane $R^2 = R × R$ where _R_ is the set of real numbers: $R^2$ is the set of all points (_x_,_y_) where _x_ and _y_ are real numbers (see the [Cartesian coordinate system](https://en.wikipedia.org/wiki/Cartesian_coordinate_system)).
The cartesian power of a set X can be defined as:
${\displaystyle X^{n}=\underbrace {X\times X\times \cdots \times X} _{n}=\{(x_{1},\ldots ,x_{n})\ |\ x_{i}\in X{\text{ for all }}i=1,\ldots ,n\}.} $
The [cardinality of a set](https://en.wikipedia.org/wiki/Cardinality) is the number of elements of the set. Cardinality of a Cartesian power set is $|S|^{n}$ where |S| is the cardinality of the set _S_ and _n_ is the power.
__We can easily use itertools again for calculating Cartesian power__. The `repeat` parameter is used as power.
```
A = {'Head','Tail'} # 2 element set
p2=list(prod(A,repeat=2)) # Power set of power 2
print("Cartesian power 2 with length {}: {}".format(len(p2),p2))
print()
p3=list(prod(A,repeat=3)) # Power set of power 3
print("Cartesian power 3 with length {}: {}".format(len(p3),p3))
```
---
## Permutations
In mathematics, the notion of permutation relates to the **act of arranging all the members of a set into some sequence or order**, or if the set is already ordered, rearranging (reordering) its elements, a process called __permuting__. The study of permutations of finite sets is a topic in the field of [combinatorics](https://en.wikipedia.org/wiki/Combinatorics).
We find the number of $k$-permutations of $A$, first by determining the set of permutations and then by calculating $\frac{|A|!}{(|A|-k)!}$. We first consider the special case of $k=|A|$, which is equivalent to finding the number of ways of ordering the elements of $A$.
```
import itertools
A = {'Red','Green','Blue'}
# Find all permutations of A
permute_all = set(itertools.permutations(A))
print("Permutations of {}".format(A))
print("-"*50)
for i in permute_all:
print(i)
print("-"*50)
print;print ("Number of permutations: ", len(permute_all))
import matplotlib.patches as mpatches
import matplotlib.pyplot as plt
from math import factorial
print("Factorial of 3:", factorial(3))
```
### Selecting _k_ items out of a set containing _n_ items and permuting
```
A = {'Red','Green','Blue','Violet'}
k=2
n = len(A)
permute_k = list(itertools.permutations(A, k))
print("{}-permutations of {}: ".format(k,A))
print("-"*50)
for i in permute_k:
print(i)
print("-"*50)
print ("Size of the permutation set = {}!/({}-{})! = {}".format(n,n,k, len(permute_k)))
factorial(4)/(factorial(4-2))
```
## Combinations
Combinatorics is an area of mathematics primarily concerned with counting, both as a means and an end in obtaining results, and certain properties of finite structures. It is closely related to many other areas of mathematics and has many applications ranging from logic to statistical physics, from evolutionary biology to computer science, etc.
Combinatorics is well known for the breadth of the problems it tackles. Combinatorial problems arise in many areas of pure mathematics, notably in algebra, [probability theory](https://en.wikipedia.org/wiki/Probability_theory), [topology](https://en.wikipedia.org/wiki/Topology), and geometry, as well as in its many application areas. Many combinatorial questions have historically been considered in isolation, giving an _ad hoc_ solution to a problem arising in some mathematical context. In the later twentieth century, however, powerful and general theoretical methods were developed, making combinatorics into an independent branch of mathematics in its own right. One of the oldest and most accessible parts of combinatorics is [graph theory](https://en.wikipedia.org/wiki/Graph_theory), which by itself has numerous natural connections to other areas. Combinatorics is used frequently in computer science to obtain formulas and estimates in the [analysis of algorithms](https://en.wikipedia.org/wiki/Analysis_of_algorithms).
We find the number of $k$-combinations of $A$, first by determining the set of combinations and then by simply calculating:
$$\frac{|A|!}{k!\times(|A|-k)!}$$
**In combination, order matters, unlike permutations.**
```
# Print all the k-combinations of A
choose_k = list(itertools.combinations(A,k))
print("%i-combinations of %s: " %(k,A))
for i in choose_k:
print(i)
print;print("Number of combinations = %i!/(%i!(%i-%i)!) = %i" %(n,k,n,k,len(choose_k) ))
```
## Putting it all together - some probability calculation examples
### Problem 1: Two dice
Two dice are rolled together. What is the probability of getting a total which is a multiple of 3?
```
n_dice = 2
dice_faces = {1,2,3,4,5,6}
# Construct the event space i.e. set of ALL POSSIBLE events
event_space = set(prod(dice_faces,repeat=n_dice))
for outcome in event_space:
print(outcome,end=', ')
# What is the set we are interested in?
favorable_outcome = []
for outcome in event_space:
x,y = outcome
if (x+y)%3==0:
favorable_outcome.append(outcome)
favorable_outcome = set(favorable_outcome)
for f_outcome in favorable_outcome:
print(f_outcome,end=', ')
prob = len(favorable_outcome)/len(event_space)
print("The probability of getting a sum which is a multiple of 3 is: ", prob)
```
### Problem 2: Five dice!
Five dice are rolled together. What is the probability of getting a total which is a multiple of 5 but not a multiple of 3?
```
n_dice = 5
dice_faces = {1,2,3,4,5,6}
# Construct the event space i.e. set of ALL POSSIBLE events
event_space = set(prod(dice_faces,repeat=n_dice))
6**5
# What is the set we are interested in?
favorable_outcome = []
for outcome in event_space:
d1,d2,d3,d4,d5 = outcome
if (d1+d2+d3+d4+d5)%5==0 and (d1+d2+d3+d4+d5)%3!=0 :
favorable_outcome.append(outcome)
favorable_outcome = set(favorable_outcome)
prob = len(favorable_outcome)/len(event_space)
print("The probability of getting a sum, which is a multiple of 5 but not a multiple of 3, is: ", prob)
```
### Problem 2 solved using set difference
```
multiple_of_5 = []
multiple_of_3 = []
for outcome in event_space:
d1,d2,d3,d4,d5 = outcome
if (d1+d2+d3+d4+d5)%5==0:
multiple_of_5.append(outcome)
if (d1+d2+d3+d4+d5)%3==0:
multiple_of_3.append(outcome)
favorable_outcome = set(multiple_of_5).difference(set(multiple_of_3))
for i in list(favorable_outcome)[:5]:
a1,a2,a3,a4,a5=i
print("{}, SUM: {}".format(i,a1+a2+a3+a4+a5))
prob = len(favorable_outcome)/len(event_space)
print("The probability of getting a sum, which is a multiple of 5 but not a multiple of 3, is: ", prob)
```
## Computing _pi_ ($\pi$) with a random dart throwing game and using probability concept
The number $\pi$ is a mathematical constant. Originally defined as the ratio of a circle's circumference to its diameter, it now has various equivalent definitions and appears in many formulas in all areas of mathematics and physics. It is approximately equal to 3.14159. It has been represented by the Greek letter __"$\pi$"__ since the mid-18th century, though it is also sometimes spelled out as "pi". It is also called Archimedes' constant.
Being an irrational number, $\pi$ cannot be expressed as a common fraction (equivalently, its decimal representation never ends and never settles into a permanently repeating pattern).
### What is the logic behind computing $\pi$ by throwing dart randomly?
Imagine a square dartboard.
Then, the dartboard with a circle drawn inside it touching all its sides.
And then, you throw darts at it. Randomly. That means some fall inside the circle, some outside. But assume that no dart falls outside the board.
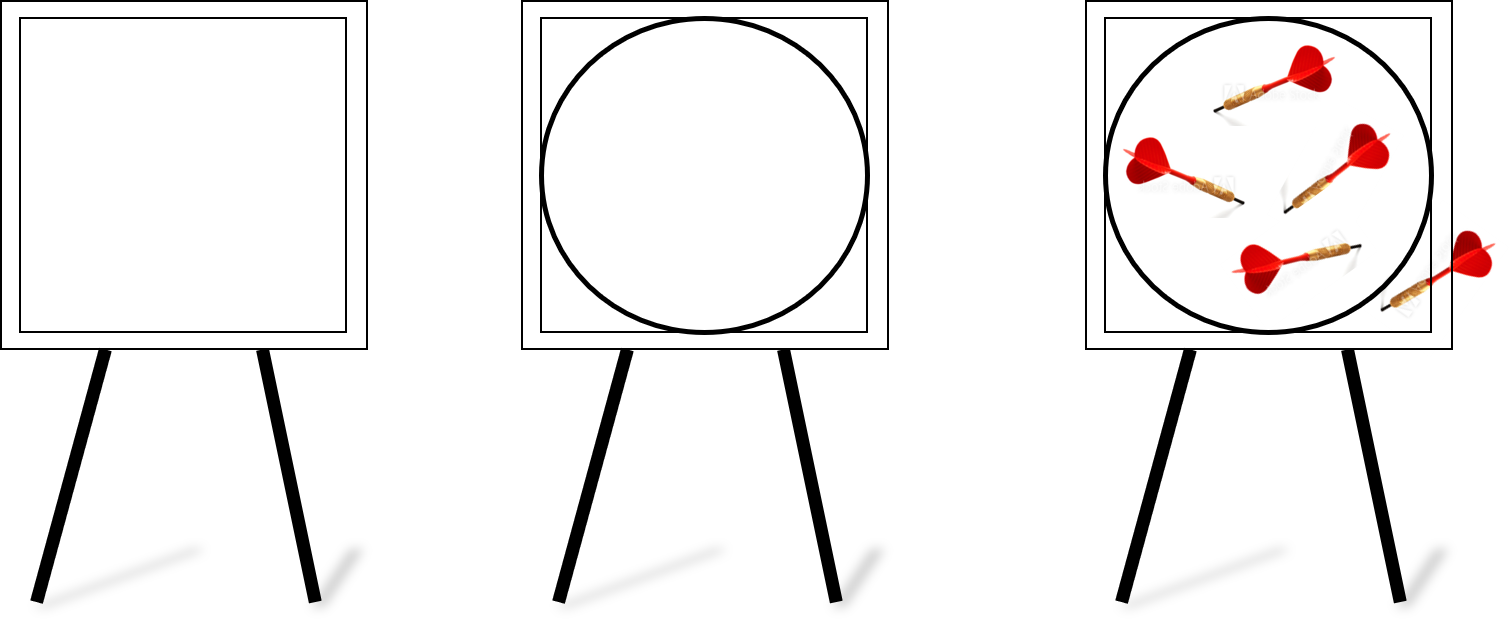
At the end of your dart throwing session,
- You count the fraction of darts that fell inside the circle of the total number of darts thrown.
- Multiply that number by 4.
- The resulting number should be pi. Or, a close approximation if you had thrown a lot of darts.
The idea is extremely simple. If you throw a large number of darts, then the **probability of a dart falling inside the circle is just the ratio of the area of the circle to that of the area of the square board**. With the help of basic mathematics, you can show that this ratio turns out to be $\frac{\pi}{4}$. So, to get $\pi$, you just multiply that number by 4.
The key here is to simulate the throwing of a lot of darts so as to make the fraction equal to the probability, an assertion valid only in the limit of a large number of trials of this random event. This comes from the [law of large number](https://en.wikipedia.org/wiki/Law_of_large_numbers) or the [frequentist definition of probability](https://en.wikipedia.org/wiki/Frequentist_probability).
See also the concept of [Buffon's Needle](https://en.wikipedia.org/wiki/Buffon%27s_needle_problem)
```
from math import pi,sqrt
import random
import matplotlib.pyplot as plt
import numpy as np
```
### Center point and the side of the square
```
# Center point
x,y = 0,0
# Side of the square
a = 2
```
### Function to simulate a random throw of a dart aiming at the square
```
def throw_dart():
"""
Simulates the randon throw of a dirt. It can land anywhere in the square (uniformly randomly)
"""
# Random final landing position of the dirt between -a/2 and +a/2 around the center point
position_x = x+a/2*(-1+2*random.random())
position_y = y+a/2*(-1+2*random.random())
return (position_x,position_y)
throw_dart()
```
### Function to determine if the dart landed inside the circle
```
def is_within_circle(x,y):
"""
Given the landing coordinate of a dirt, determines if it fell inside the circle
"""
# Side of the square
a = 2
distance_from_center = sqrt(x**2+y**2)
if distance_from_center < a/2:
return True
else:
return False
is_within_circle(1.9,1.9)
is_within_circle(0.2,-0.6)
```
### Now, throw a few darts
```
n_throws = 10
count_inside_circle=0
for i in range(n_throws):
r1,r2=throw_dart()
if is_within_circle(r1,r2):
count_inside_circle+=1
```
### Compute the ratio of `count_inside_circle` and `n_throws`
```
ratio = count_inside_circle/n_throws
```
### Is it approximately equal to $\frac{\pi}{4}$?
```
print(4*ratio)
```
### Not exactly. Let's try with a lot more darts!
```
n_throws = 10000
count_inside_circle=0
for i in range(n_throws):
r1,r2=throw_dart()
if is_within_circle(r1,r2):
count_inside_circle+=1
ratio = count_inside_circle/n_throws
print(4*ratio)
```
### Let's functionalize this process and run a number of times
```
def compute_pi_throwing_dart(n_throws):
"""
Computes pi by throwing a bunch of darts at the square
"""
n_throws = n_throws
count_inside_circle=0
for i in range(n_throws):
r1,r2=throw_dart()
if is_within_circle(r1,r2):
count_inside_circle+=1
result = 4*(count_inside_circle/n_throws)
return result
```
### Now let us run this experiment a few times and see what happens.
```
n_exp=[]
pi_exp=[]
n = [int(10**(0.5*i)) for i in range(1,15)]
for i in n:
p = compute_pi_throwing_dart(i)
pi_exp.append(p)
n_exp.append(i)
print("Computed value of pi by throwing {} darts is: {}".format(i,p))
plt.figure(figsize=(8,5))
plt.title("Computing pi with \nincreasing number of random throws",fontsize=20)
plt.semilogx(n_exp, pi_exp,c='k',marker='o',lw=3)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Number of random throws",fontsize=15)
plt.ylabel("Computed value of pi",fontsize=15)
plt.hlines(y=3.14159,xmin=1,xmax=1e7,linestyle='--')
plt.text(x=10,y=3.05,s="Value of pi",fontsize=17)
plt.grid(True)
plt.show()
```
|
github_jupyter
|
# Directly with curly braces
Set1 = {1,2}
print (Set1)
print(type(Set1))
my_list=[1,2,3,4]
my_set_from_list = set(my_list)
print(my_set_from_list)
my_set = set([1,3,5])
print("Here is my set:",my_set)
print("1 is in the set:",1 in my_set)
print("2 is in the set:",2 in my_set)
print("4 is NOT in the set:",4 not in my_set)
Univ = set([x for x in range(11)])
Super = set([x for x in range(11) if x%2==0])
disj = set([x for x in range(11) if x%2==1])
Sub = set([4,6])
Null = set([x for x in range(11) if x>10])
print("Universal set (all the positive integers up to 10):",Univ)
print("All the even positive integers up to 10:",Super)
print("All the odd positive integers up to 10:",disj)
print("Set of 2 elements, 4 and 6:",Sub)
print("A null set:", Null)
print('Is "Super" a superset of "Sub"?',Super.issuperset(Sub))
print('Is "Super" a subset of "Univ"?',Super.issubset(Univ))
print('Is "Sub" a superset of "Super"?',Sub.issuperset(Super))
print('Is "Super" disjoint with "disj"?',Sub.isdisjoint(disj))
S1 = {1,2}
S2 = {2,2,1,1,2}
print ("S1 and S2 are equal because order or repetition of elements do not matter for sets\nS1==S2:", S1==S2)
S1 = {1,2,3,4,5,6}
S2 = {1,2,3,4,0,6}
print ("S1 and S2 are NOT equal because at least one element is different\nS1==S2:", S1==S2)
# Define a set using list comprehension
S1 = set([x for x in range(1,11) if x%3==0])
print("S1:", S1)
S2 = set([x for x in range(1,7)])
print("S2:", S2)
# Both intersection method or & can be used
S_intersection = S1.intersection(S2)
print("Intersection of S1 and S2:", S_intersection)
S_intersection = S1 & S2
print("Intersection of S1 and S2:", S_intersection)
S3 = set([x for x in range(6,10)])
print("S3:", S3)
S1_S2_S3 = S1.intersection(S2).intersection(S3)
print("Intersection of S1, S2, and S3:", S1_S2_S3)
# Both union method or | can be used
S1 = set([x for x in range(1,11) if x%3==0])
print("S1:", S1)
S2 = set([x for x in range(1,5)])
print("S2:", S2)
S_union = S1.union(S2)
print("Union of S1 and S2:", S_union)
S_union = S1 | S2
print("Union of S1 and S2:", S_union)
S=set([x for x in range (21) if x%2==0])
print ("S is the set of even numbers between 0 and 20:", S)
S_complement = set([x for x in range (21) if x%2!=0])
print ("S_complement is the set of odd numbers between 0 and 20:", S_complement)
print ("Is the union of S and S_complement equal to all numbers between 0 and 20?",
S.union(S_complement)==set([x for x in range (21)]))
S1 = set([x for x in range(31) if x%3==0])
print ("Set S1:", S1)
S2 = set([x for x in range(31) if x%5==0])
print ("Set S2:", S2)
S_difference = S2-S1
print("Difference of S1 and S2 i.e. S2\S1:", S_difference)
S_difference = S1.difference(S2)
print("Difference of S2 and S1 i.e. S1\S2:", S_difference)
print("S1",S1)
print("S2",S2)
print("Symmetric difference", S1^S2)
print("Symmetric difference", S2.symmetric_difference(S1))
A = set(['a','b','c'])
S = {1,2,3}
def cartesian_product(S1,S2):
result = set()
for i in S1:
for j in S2:
result.add(tuple([i,j]))
return (result)
C = cartesian_product(A,S)
print("Cartesian product of A and S\n{} X {}:{}".format(A,S,C))
print("Length of the Cartesian product set:",len(C))
from itertools import product as prod
A = set([x for x in range(1,7)])
B = set([x for x in range(1,7)])
p=list(prod(A,B))
print("A is set of all possible throws of a dice:",A)
print("B is set of all possible throws of a dice:",B)
print ("\nProduct of A and B is the all possible combinations of A and B thrown together:\n",p)
A = {'Head','Tail'} # 2 element set
p2=list(prod(A,repeat=2)) # Power set of power 2
print("Cartesian power 2 with length {}: {}".format(len(p2),p2))
print()
p3=list(prod(A,repeat=3)) # Power set of power 3
print("Cartesian power 3 with length {}: {}".format(len(p3),p3))
import itertools
A = {'Red','Green','Blue'}
# Find all permutations of A
permute_all = set(itertools.permutations(A))
print("Permutations of {}".format(A))
print("-"*50)
for i in permute_all:
print(i)
print("-"*50)
print;print ("Number of permutations: ", len(permute_all))
import matplotlib.patches as mpatches
import matplotlib.pyplot as plt
from math import factorial
print("Factorial of 3:", factorial(3))
A = {'Red','Green','Blue','Violet'}
k=2
n = len(A)
permute_k = list(itertools.permutations(A, k))
print("{}-permutations of {}: ".format(k,A))
print("-"*50)
for i in permute_k:
print(i)
print("-"*50)
print ("Size of the permutation set = {}!/({}-{})! = {}".format(n,n,k, len(permute_k)))
factorial(4)/(factorial(4-2))
# Print all the k-combinations of A
choose_k = list(itertools.combinations(A,k))
print("%i-combinations of %s: " %(k,A))
for i in choose_k:
print(i)
print;print("Number of combinations = %i!/(%i!(%i-%i)!) = %i" %(n,k,n,k,len(choose_k) ))
n_dice = 2
dice_faces = {1,2,3,4,5,6}
# Construct the event space i.e. set of ALL POSSIBLE events
event_space = set(prod(dice_faces,repeat=n_dice))
for outcome in event_space:
print(outcome,end=', ')
# What is the set we are interested in?
favorable_outcome = []
for outcome in event_space:
x,y = outcome
if (x+y)%3==0:
favorable_outcome.append(outcome)
favorable_outcome = set(favorable_outcome)
for f_outcome in favorable_outcome:
print(f_outcome,end=', ')
prob = len(favorable_outcome)/len(event_space)
print("The probability of getting a sum which is a multiple of 3 is: ", prob)
n_dice = 5
dice_faces = {1,2,3,4,5,6}
# Construct the event space i.e. set of ALL POSSIBLE events
event_space = set(prod(dice_faces,repeat=n_dice))
6**5
# What is the set we are interested in?
favorable_outcome = []
for outcome in event_space:
d1,d2,d3,d4,d5 = outcome
if (d1+d2+d3+d4+d5)%5==0 and (d1+d2+d3+d4+d5)%3!=0 :
favorable_outcome.append(outcome)
favorable_outcome = set(favorable_outcome)
prob = len(favorable_outcome)/len(event_space)
print("The probability of getting a sum, which is a multiple of 5 but not a multiple of 3, is: ", prob)
multiple_of_5 = []
multiple_of_3 = []
for outcome in event_space:
d1,d2,d3,d4,d5 = outcome
if (d1+d2+d3+d4+d5)%5==0:
multiple_of_5.append(outcome)
if (d1+d2+d3+d4+d5)%3==0:
multiple_of_3.append(outcome)
favorable_outcome = set(multiple_of_5).difference(set(multiple_of_3))
for i in list(favorable_outcome)[:5]:
a1,a2,a3,a4,a5=i
print("{}, SUM: {}".format(i,a1+a2+a3+a4+a5))
prob = len(favorable_outcome)/len(event_space)
print("The probability of getting a sum, which is a multiple of 5 but not a multiple of 3, is: ", prob)
from math import pi,sqrt
import random
import matplotlib.pyplot as plt
import numpy as np
# Center point
x,y = 0,0
# Side of the square
a = 2
def throw_dart():
"""
Simulates the randon throw of a dirt. It can land anywhere in the square (uniformly randomly)
"""
# Random final landing position of the dirt between -a/2 and +a/2 around the center point
position_x = x+a/2*(-1+2*random.random())
position_y = y+a/2*(-1+2*random.random())
return (position_x,position_y)
throw_dart()
def is_within_circle(x,y):
"""
Given the landing coordinate of a dirt, determines if it fell inside the circle
"""
# Side of the square
a = 2
distance_from_center = sqrt(x**2+y**2)
if distance_from_center < a/2:
return True
else:
return False
is_within_circle(1.9,1.9)
is_within_circle(0.2,-0.6)
n_throws = 10
count_inside_circle=0
for i in range(n_throws):
r1,r2=throw_dart()
if is_within_circle(r1,r2):
count_inside_circle+=1
ratio = count_inside_circle/n_throws
print(4*ratio)
n_throws = 10000
count_inside_circle=0
for i in range(n_throws):
r1,r2=throw_dart()
if is_within_circle(r1,r2):
count_inside_circle+=1
ratio = count_inside_circle/n_throws
print(4*ratio)
def compute_pi_throwing_dart(n_throws):
"""
Computes pi by throwing a bunch of darts at the square
"""
n_throws = n_throws
count_inside_circle=0
for i in range(n_throws):
r1,r2=throw_dart()
if is_within_circle(r1,r2):
count_inside_circle+=1
result = 4*(count_inside_circle/n_throws)
return result
n_exp=[]
pi_exp=[]
n = [int(10**(0.5*i)) for i in range(1,15)]
for i in n:
p = compute_pi_throwing_dart(i)
pi_exp.append(p)
n_exp.append(i)
print("Computed value of pi by throwing {} darts is: {}".format(i,p))
plt.figure(figsize=(8,5))
plt.title("Computing pi with \nincreasing number of random throws",fontsize=20)
plt.semilogx(n_exp, pi_exp,c='k',marker='o',lw=3)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Number of random throws",fontsize=15)
plt.ylabel("Computed value of pi",fontsize=15)
plt.hlines(y=3.14159,xmin=1,xmax=1e7,linestyle='--')
plt.text(x=10,y=3.05,s="Value of pi",fontsize=17)
plt.grid(True)
plt.show()
| 0.298798 | 0.960621 |
# Amazon SageMaker Object Detection using the augmented manifest file format
1. [Introduction](#Introduction)
2. [Setup](#Setup)
3. [Specifying input Dataset](#Specifying-input-Dataset)
4. [Training](#Training)
## Introduction
Object detection is the process of identifying and localizing objects in an image. A typical object detection solution takes in an image as input and provides a bounding box on the image where an object of interest is, along with identifying what object the box encapsulates. But before we have this solution, we need to process a training dataset, create and setup a training job for the algorithm so that the aglorithm can learn about the dataset and then host the algorithm as an endpoint, to which we can supply the query image.
This notebook focuses on using the built-in SageMaker Single Shot multibox Detector ([SSD](https://arxiv.org/abs/1512.02325)) object detection algorithm to train model on your custom dataset. For dataset prepration or using the model for inference, please see other scripts in [this folder](./)
## Setup
To train the Object Detection algorithm on Amazon SageMaker, we need to setup and authenticate the use of AWS services. To begin with we need an AWS account role with SageMaker access. This role is used to give SageMaker access to your data in S3. In this example, we will use the same role that was used to start this SageMaker notebook.
```
%%time
import sagemaker
import boto3
from sagemaker import get_execution_role
role = get_execution_role()
print(role)
```
We also need the S3 bucket that has the training manifests and will be used to store the tranied model artifacts.
```
bucket = '<please replace with your s3 bucket name>'
prefix = 'demo'
```
## Specifying input Dataset
This notebook assumes you already have prepared two [Augmented Manifest Files](https://docs.aws.amazon.com/sagemaker/latest/dg/augmented-manifest.html) as training and validation input data for the object detection model.
There are many advantages to using **augmented manifest files** for your training input
* No format conversion is required if you are using SageMaker Ground Truth to generate the data labels
* Unlike the traditional approach of providing paths to the input images separately from its labels, augmented manifest file already combines both into one entry for each input image, reducing complexity in algorithm code for matching each image with labels. (Read this [blog post](https://aws.amazon.com/blogs/machine-learning/easily-train-models-using-datasets-labeled-by-amazon-sagemaker-ground-truth/) for more explanation.)
* When splitting your dataset for train/validation/test, you don't need to rearrange and re-upload image files to different s3 prefixes for train vs validation. Once you upload your image files to S3, you never need to move it again. You can just place pointers to these images in your augmented manifest file for training and validation. More on the train/validation data split in this post later.
* When using augmented manifest file, the training input images is loaded on to the training instance in *Pipe mode,* which means the input data is streamed directly to the training algorithm while it is running (vs. File mode, where all input files need to be downloaded to disk before the training starts). This results in faster training performance and less disk resource utilization. Read more in this [blog post](https://aws.amazon.com/blogs/machine-learning/accelerate-model-training-using-faster-pipe-mode-on-amazon-sagemaker/) on the benefits of pipe mode.
```
train_data_prefix = "demo"
# below uses the training data after augmentation
s3_train_data= "s3://{}/{}/all_augmented.json".format(bucket, train_data_prefix)
# uncomment below to use the non-augmented input
# s3_train_data= "s3://{}/training-manifest/{}/train.manifest".format(bucket, train_data_prefix)
s3_validation_data = "s3://{}/training-manifest/{}/validation.manifest".format(bucket, train_data_prefix)
print("Train data: {}".format(s3_train_data) )
print("Validation data: {}".format(s3_validation_data) )
train_input = {
"ChannelName": "train",
"DataSource": {
"S3DataSource": {
"S3DataType": "AugmentedManifestFile",
"S3Uri": s3_train_data,
"S3DataDistributionType": "FullyReplicated",
# This must correspond to the JSON field names in your augmented manifest.
"AttributeNames": ['source-ref', 'bb']
}
},
"ContentType": "application/x-recordio",
"RecordWrapperType": "RecordIO",
"CompressionType": "None"
}
validation_input = {
"ChannelName": "validation",
"DataSource": {
"S3DataSource": {
"S3DataType": "AugmentedManifestFile",
"S3Uri": s3_validation_data,
"S3DataDistributionType": "FullyReplicated",
# This must correspond to the JSON field names in your augmented manifest.
"AttributeNames": ['source-ref', 'bb']
}
},
"ContentType": "application/x-recordio",
"RecordWrapperType": "RecordIO",
"CompressionType": "None"
}
```
Below code computes the number of training samples, required in the training job request.
```
import json
import os
def read_manifest_file(file_path):
with open(file_path, 'r') as f:
output = [json.loads(line.strip()) for line in f.readlines()]
return output
!aws s3 cp $s3_train_data .
train_data = read_manifest_file(os.path.split(s3_train_data)[1])
num_training_samples = len(train_data)
num_training_samples
s3_output_path = 's3://{}/{}/output'.format(bucket, prefix)
s3_output_path
```
## Training
Now that we are done with all the setup that is needed, we are ready to train our object detector.
```
from sagemaker.amazon.amazon_estimator import get_image_uri
# This retrieves a docker container with the built in object detection SSD model.
training_image = sagemaker.amazon.amazon_estimator.get_image_uri(boto3.Session().region_name, 'object-detection', repo_version='latest')
print (training_image)
```
Create a unique job name
```
import time
job_name_prefix = 'od-demo'
timestamp = time.strftime('-%Y-%m-%d-%H-%M-%S', time.gmtime())
model_job_name = job_name_prefix + timestamp
model_job_name
```
The object detection algorithm at its core is the [Single-Shot Multi-Box detection algorithm (SSD)](https://arxiv.org/abs/1512.02325). This algorithm uses a `base_network`, which is typically a [VGG](https://arxiv.org/abs/1409.1556) or a [ResNet](https://arxiv.org/abs/1512.03385). (resnet is typically faster so for edge inferences, I'd recommend using this base network). The Amazon SageMaker object detection algorithm supports VGG-16 and ResNet-50 now. It also has a lot of options for hyperparameters that help configure the training job. The next step in our training, is to setup these hyperparameters and data channels for training the model. See the SageMaker Object Detection [documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/object-detection.html) for more details on the hyperparameters.
To figure out which works best for your data, run a hyperparameter tuning job. There's some example notebooks at [https://github.com/awslabs/amazon-sagemaker-examples](https://github.com/awslabs/amazon-sagemaker-examples) that you can use for reference.
```
# This is where transfer learning happens. We use the pre-trained model and nuke the output layer by specifying
# the num_classes value. You can also run a hyperparameter tuning job to figure out which values work the best.
hyperparams = {
"base_network": 'resnet-50',
"use_pretrained_model": "1",
"num_classes": "2",
"mini_batch_size": "30",
"epochs": "30",
"learning_rate": "0.001",
"lr_scheduler_step": "10,20",
"lr_scheduler_factor": "0.25",
"optimizer": "sgd",
"momentum": "0.9",
"weight_decay": "0.0005",
"overlap_threshold": "0.5",
"nms_threshold": "0.45",
"image_shape": "512",
"label_width": "150",
"num_training_samples": str(num_training_samples)
}
```
Now that the hyperparameters are set up, we configure the rest of the training job parameters
```
training_params = \
{
"AlgorithmSpecification": {
"TrainingImage": training_image,
"TrainingInputMode": "Pipe"
},
"RoleArn": role,
"OutputDataConfig": {
"S3OutputPath": s3_output_path
},
"ResourceConfig": {
"InstanceCount": 1,
"InstanceType": "ml.p3.8xlarge",
"VolumeSizeInGB": 200
},
"TrainingJobName": model_job_name,
"HyperParameters": hyperparams,
"StoppingCondition": {
"MaxRuntimeInSeconds": 86400
},
"InputDataConfig": [
train_input,
validation_input
]
}
```
Now we create the SageMaker training job.
```
client = boto3.client(service_name='sagemaker')
client.create_training_job(**training_params)
# Confirm that the training job has started
status = client.describe_training_job(TrainingJobName=model_job_name)['TrainingJobStatus']
print('Training job current status: {}'.format(status))
```
To check the progess of the training job, you can repeatedly evaluate the following cell. When the training job status reads 'Completed', move on to the next part of the tutorial.
```
client = boto3.client(service_name='sagemaker')
print("Training job status: ", client.describe_training_job(TrainingJobName=model_job_name)['TrainingJobStatus'])
print("Secondary status: ", client.describe_training_job(TrainingJobName=model_job_name)['SecondaryStatus'])
```
# Next step
Once the training job completes, move on to the [next notebook](./03_local_inference_post_training.ipynb) to convert the trained model to a deployable format and run local inference
|
github_jupyter
|
%%time
import sagemaker
import boto3
from sagemaker import get_execution_role
role = get_execution_role()
print(role)
bucket = '<please replace with your s3 bucket name>'
prefix = 'demo'
train_data_prefix = "demo"
# below uses the training data after augmentation
s3_train_data= "s3://{}/{}/all_augmented.json".format(bucket, train_data_prefix)
# uncomment below to use the non-augmented input
# s3_train_data= "s3://{}/training-manifest/{}/train.manifest".format(bucket, train_data_prefix)
s3_validation_data = "s3://{}/training-manifest/{}/validation.manifest".format(bucket, train_data_prefix)
print("Train data: {}".format(s3_train_data) )
print("Validation data: {}".format(s3_validation_data) )
train_input = {
"ChannelName": "train",
"DataSource": {
"S3DataSource": {
"S3DataType": "AugmentedManifestFile",
"S3Uri": s3_train_data,
"S3DataDistributionType": "FullyReplicated",
# This must correspond to the JSON field names in your augmented manifest.
"AttributeNames": ['source-ref', 'bb']
}
},
"ContentType": "application/x-recordio",
"RecordWrapperType": "RecordIO",
"CompressionType": "None"
}
validation_input = {
"ChannelName": "validation",
"DataSource": {
"S3DataSource": {
"S3DataType": "AugmentedManifestFile",
"S3Uri": s3_validation_data,
"S3DataDistributionType": "FullyReplicated",
# This must correspond to the JSON field names in your augmented manifest.
"AttributeNames": ['source-ref', 'bb']
}
},
"ContentType": "application/x-recordio",
"RecordWrapperType": "RecordIO",
"CompressionType": "None"
}
import json
import os
def read_manifest_file(file_path):
with open(file_path, 'r') as f:
output = [json.loads(line.strip()) for line in f.readlines()]
return output
!aws s3 cp $s3_train_data .
train_data = read_manifest_file(os.path.split(s3_train_data)[1])
num_training_samples = len(train_data)
num_training_samples
s3_output_path = 's3://{}/{}/output'.format(bucket, prefix)
s3_output_path
from sagemaker.amazon.amazon_estimator import get_image_uri
# This retrieves a docker container with the built in object detection SSD model.
training_image = sagemaker.amazon.amazon_estimator.get_image_uri(boto3.Session().region_name, 'object-detection', repo_version='latest')
print (training_image)
import time
job_name_prefix = 'od-demo'
timestamp = time.strftime('-%Y-%m-%d-%H-%M-%S', time.gmtime())
model_job_name = job_name_prefix + timestamp
model_job_name
# This is where transfer learning happens. We use the pre-trained model and nuke the output layer by specifying
# the num_classes value. You can also run a hyperparameter tuning job to figure out which values work the best.
hyperparams = {
"base_network": 'resnet-50',
"use_pretrained_model": "1",
"num_classes": "2",
"mini_batch_size": "30",
"epochs": "30",
"learning_rate": "0.001",
"lr_scheduler_step": "10,20",
"lr_scheduler_factor": "0.25",
"optimizer": "sgd",
"momentum": "0.9",
"weight_decay": "0.0005",
"overlap_threshold": "0.5",
"nms_threshold": "0.45",
"image_shape": "512",
"label_width": "150",
"num_training_samples": str(num_training_samples)
}
training_params = \
{
"AlgorithmSpecification": {
"TrainingImage": training_image,
"TrainingInputMode": "Pipe"
},
"RoleArn": role,
"OutputDataConfig": {
"S3OutputPath": s3_output_path
},
"ResourceConfig": {
"InstanceCount": 1,
"InstanceType": "ml.p3.8xlarge",
"VolumeSizeInGB": 200
},
"TrainingJobName": model_job_name,
"HyperParameters": hyperparams,
"StoppingCondition": {
"MaxRuntimeInSeconds": 86400
},
"InputDataConfig": [
train_input,
validation_input
]
}
client = boto3.client(service_name='sagemaker')
client.create_training_job(**training_params)
# Confirm that the training job has started
status = client.describe_training_job(TrainingJobName=model_job_name)['TrainingJobStatus']
print('Training job current status: {}'.format(status))
client = boto3.client(service_name='sagemaker')
print("Training job status: ", client.describe_training_job(TrainingJobName=model_job_name)['TrainingJobStatus'])
print("Secondary status: ", client.describe_training_job(TrainingJobName=model_job_name)['SecondaryStatus'])
| 0.289573 | 0.981113 |
# Objective
Investigate the prevalence of workers with outlier pairwise scores. Do datasets consistently have workers with outlier pairwise scores?
For each worker, score = sum(pair scores of that worker).
For worker_A and worker_B, pair_score = [(avg NND worker_A -> worker_B) + (avg NND worker_B -> worker_A)] / 2.
```
from SpotAnnotationAnalysis import SpotAnnotationAnalysis
from BaseAnnotation import BaseAnnotation
from QuantiusAnnotation import QuantiusAnnotation
worker_marker_size = 8
cluster_marker_size = 40
bigger_window_size = False
img_height = 300
correctness_threshold = 4
clustering_params = ['AffinityPropagation', -350]
```
# Takeaways
Datasets consistently have workers with outlier pairwise scores.
In some datasets in this batch, e.g. MAX_ISP_300_1_nspots100_spot_sig1.75_snr20_2.5, there is less of clear cutoff between outliers and other workers. There tends to be a clearer cutoff between outliers and other workers when there are fewer spots.
In all datasets in this batch, the majority of worker scores cluster low.
# Plots
Grouped by:
- background
- number of spots
- mean SNR
## Background: Tissue
```
json_filename = 'SynthTests_tissue.json'
gen_date = '20180719'
bg_type = 'tissue'
```
## Tissue, 50 spots
```
img_names = ['MAX_ISP_300_1_nspots50_spot_sig1.75_snr5_2.5',
'MAX_ISP_300_1_nspots50_spot_sig1.75_snr10_2.5',
'MAX_ISP_300_1_nspots50_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
```
## Tissue, 100 spots
```
img_names = ['MAX_ISP_300_1_nspots100_spot_sig1.75_snr5_2.5',
'MAX_ISP_300_1_nspots100_spot_sig1.75_snr10_2.5',
'MAX_ISP_300_1_nspots100_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
```
## Tissue, 150 spots
```
img_names = ['MAX_ISP_300_1_nspots150_spot_sig1.75_snr5_2.5',
'MAX_ISP_300_1_nspots150_spot_sig1.75_snr10_2.5',
'MAX_ISP_300_1_nspots150_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
```
# Background: Cells
```
json_filename = 'SynthData_cells.json'
gen_date = '20180719'
bg_type = 'cells'
```
## Cells, 50 spots
```
img_names = ['MAX_C3-ISP_300_1_nspots50_spot_sig1.75_snr5_2.5',
'MAX_C3-ISP_300_1_nspots50_spot_sig1.75_snr10_2.5',
'MAX_C3-ISP_300_1_nspots50_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
```
## Cells, 100 spots
```
img_names = ['MAX_C3-ISP_300_1_nspots100_spot_sig1.75_snr5_2.5',
'MAX_C3-ISP_300_1_nspots100_spot_sig1.75_snr10_2.5',
'MAX_C3-ISP_300_1_nspots100_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
```
## Cells, 150 spots
```
img_names = ['MAX_C3-ISP_300_1_nspots150_spot_sig1.75_snr5_2.5',
'MAX_C3-ISP_300_1_nspots150_spot_sig1.75_snr10_2.5',
'MAX_C3-ISP_300_1_nspots150_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
```
|
github_jupyter
|
from SpotAnnotationAnalysis import SpotAnnotationAnalysis
from BaseAnnotation import BaseAnnotation
from QuantiusAnnotation import QuantiusAnnotation
worker_marker_size = 8
cluster_marker_size = 40
bigger_window_size = False
img_height = 300
correctness_threshold = 4
clustering_params = ['AffinityPropagation', -350]
json_filename = 'SynthTests_tissue.json'
gen_date = '20180719'
bg_type = 'tissue'
img_names = ['MAX_ISP_300_1_nspots50_spot_sig1.75_snr5_2.5',
'MAX_ISP_300_1_nspots50_spot_sig1.75_snr10_2.5',
'MAX_ISP_300_1_nspots50_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
img_names = ['MAX_ISP_300_1_nspots100_spot_sig1.75_snr5_2.5',
'MAX_ISP_300_1_nspots100_spot_sig1.75_snr10_2.5',
'MAX_ISP_300_1_nspots100_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
img_names = ['MAX_ISP_300_1_nspots150_spot_sig1.75_snr5_2.5',
'MAX_ISP_300_1_nspots150_spot_sig1.75_snr10_2.5',
'MAX_ISP_300_1_nspots150_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
json_filename = 'SynthData_cells.json'
gen_date = '20180719'
bg_type = 'cells'
img_names = ['MAX_C3-ISP_300_1_nspots50_spot_sig1.75_snr5_2.5',
'MAX_C3-ISP_300_1_nspots50_spot_sig1.75_snr10_2.5',
'MAX_C3-ISP_300_1_nspots50_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
img_names = ['MAX_C3-ISP_300_1_nspots100_spot_sig1.75_snr5_2.5',
'MAX_C3-ISP_300_1_nspots100_spot_sig1.75_snr10_2.5',
'MAX_C3-ISP_300_1_nspots100_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
img_names = ['MAX_C3-ISP_300_1_nspots150_spot_sig1.75_snr5_2.5',
'MAX_C3-ISP_300_1_nspots150_spot_sig1.75_snr10_2.5',
'MAX_C3-ISP_300_1_nspots150_spot_sig1.75_snr20_2.5']
for img_name in img_names:
img_filename = img_name+'spot_img.png'
img_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_images/'+bg_type+'/'+img_name+'spot_img.png'
csv_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/spot_data/'+bg_type+'/'+img_name+'_coord_snr_list.csv'
json_filepath = '/Users/jenny.vo-phamhi/Documents/FISH-annotation/Annotation/gen_'+gen_date+'/'+json_filename
ba = QuantiusAnnotation(json_filepath)
sa = SpotAnnotationAnalysis(ba)
anno_all = ba.df()
anno_one_snr = ba.slice_by_image(anno_all, img_filename)
plot_title = img_name
sa.plot_worker_pairwise_scores_hist(anno_one_snr, bigger_window_size, plot_title)
| 0.236428 | 0.748421 |
```
%matplotlib inline
import os
import sys
import netCDF4
import numpy as np
from geophys_utils import NetCDFPointUtils, get_spatial_ref_from_wkt
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.io.img_tiles as cimgt
from pprint import pprint
#print(sys.version)
#pprint(dict(os.environ))
def rescale_array(input_np_array, new_range_min=0, new_range_max=1):
old_min = input_np_array.min()
old_range = input_np_array.max() - old_min
new_range = new_range_max - new_range_min
scaled_np_array = ((input_np_array - old_min) / old_range * new_range) + new_range_min
return scaled_np_array
def plot_survey_points(netcdf_path, variable_to_map, colour_scheme='binary'):
nc = netCDF4.Dataset(netcdf_path)
netcdf_point_utils = NetCDFPointUtils(nc)
utm_wkt, utm_coords = netcdf_point_utils.utm_coords(netcdf_point_utils.xycoords[:])
utm_zone = get_spatial_ref_from_wkt(utm_wkt).GetUTMZone() # -ve for Southern Hemisphere
southern_hemisphere = (utm_zone < 0)
utm_zone = abs(utm_zone)
projection = ccrs.UTM(zone=utm_zone, southern_hemisphere=southern_hemisphere)
#print(nc.variables)
variable = nc.variables[variable_to_map][:]
colour_array = rescale_array(variable, 0, 1)
#map_image = cimgt.OSM()# http://developer.mapquest.com/web/products/open/map for terms of use
#map_image = cimgt.StamenTerrain() # http://maps.stamen.com/
map_image = cimgt.QuadtreeTiles()
fig = plt.figure(figsize=(30,20))
ax = fig.add_subplot(1, 1, 1, projection=projection)
ax.set_title("Point Gravity Survey - " + str(nc.getncattr('title')))
ax.add_image(map_image, 10)
# set the x and y axis tick values
x_min = np.min(utm_coords[:,0])
x_max = np.max(utm_coords[:,0])
y_min = np.min(utm_coords[:,1])
y_max = np.max(utm_coords[:,1])
range_x = x_max - x_min
range_y = y_max - y_min
ax.set_xticks([x_min, range_x / 2 + x_min, x_max])
ax.set_yticks([y_min, range_y / 2 + y_min, y_max])
# set the x and y axis labels
ax.set_xlabel("Eastings (m)", rotation=0, labelpad=20)
ax.set_ylabel("Northings (m)", rotation=90, labelpad=20)
# See link for possible colourmap schemes: https://matplotlib.org/examples/color/colormaps_reference.html
cm = plt.cm.get_cmap(colour_scheme)
# build a scatter plot of the specified data, define marker, spatial reference system, and the chosen colour map type
sc = ax.scatter(utm_coords[:,0],
utm_coords[:,1],
marker='o',
c=colour_array,
s=4,
alpha=0.9,
transform=projection,
cmap=cm
)
# set the colour bar ticks and labels
cb = plt.colorbar(sc, ticks=[0, 1])
cb.ax.set_yticklabels([str(np.min(variable)), str(np.max(variable))]) # vertically oriented colorbar
cb.set_label("Free Air Anomaly (um/s^2)")
plt.show()
nc_path = 'http://dapds00.nci.org.au/thredds/dodsC/uc0/rr2_dev/axi547/ground_gravity/point_datasets/194701.nc'
plot_survey_points(nc_path, 'Freeair', 'gist_heat')
```
|
github_jupyter
|
%matplotlib inline
import os
import sys
import netCDF4
import numpy as np
from geophys_utils import NetCDFPointUtils, get_spatial_ref_from_wkt
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.io.img_tiles as cimgt
from pprint import pprint
#print(sys.version)
#pprint(dict(os.environ))
def rescale_array(input_np_array, new_range_min=0, new_range_max=1):
old_min = input_np_array.min()
old_range = input_np_array.max() - old_min
new_range = new_range_max - new_range_min
scaled_np_array = ((input_np_array - old_min) / old_range * new_range) + new_range_min
return scaled_np_array
def plot_survey_points(netcdf_path, variable_to_map, colour_scheme='binary'):
nc = netCDF4.Dataset(netcdf_path)
netcdf_point_utils = NetCDFPointUtils(nc)
utm_wkt, utm_coords = netcdf_point_utils.utm_coords(netcdf_point_utils.xycoords[:])
utm_zone = get_spatial_ref_from_wkt(utm_wkt).GetUTMZone() # -ve for Southern Hemisphere
southern_hemisphere = (utm_zone < 0)
utm_zone = abs(utm_zone)
projection = ccrs.UTM(zone=utm_zone, southern_hemisphere=southern_hemisphere)
#print(nc.variables)
variable = nc.variables[variable_to_map][:]
colour_array = rescale_array(variable, 0, 1)
#map_image = cimgt.OSM()# http://developer.mapquest.com/web/products/open/map for terms of use
#map_image = cimgt.StamenTerrain() # http://maps.stamen.com/
map_image = cimgt.QuadtreeTiles()
fig = plt.figure(figsize=(30,20))
ax = fig.add_subplot(1, 1, 1, projection=projection)
ax.set_title("Point Gravity Survey - " + str(nc.getncattr('title')))
ax.add_image(map_image, 10)
# set the x and y axis tick values
x_min = np.min(utm_coords[:,0])
x_max = np.max(utm_coords[:,0])
y_min = np.min(utm_coords[:,1])
y_max = np.max(utm_coords[:,1])
range_x = x_max - x_min
range_y = y_max - y_min
ax.set_xticks([x_min, range_x / 2 + x_min, x_max])
ax.set_yticks([y_min, range_y / 2 + y_min, y_max])
# set the x and y axis labels
ax.set_xlabel("Eastings (m)", rotation=0, labelpad=20)
ax.set_ylabel("Northings (m)", rotation=90, labelpad=20)
# See link for possible colourmap schemes: https://matplotlib.org/examples/color/colormaps_reference.html
cm = plt.cm.get_cmap(colour_scheme)
# build a scatter plot of the specified data, define marker, spatial reference system, and the chosen colour map type
sc = ax.scatter(utm_coords[:,0],
utm_coords[:,1],
marker='o',
c=colour_array,
s=4,
alpha=0.9,
transform=projection,
cmap=cm
)
# set the colour bar ticks and labels
cb = plt.colorbar(sc, ticks=[0, 1])
cb.ax.set_yticklabels([str(np.min(variable)), str(np.max(variable))]) # vertically oriented colorbar
cb.set_label("Free Air Anomaly (um/s^2)")
plt.show()
nc_path = 'http://dapds00.nci.org.au/thredds/dodsC/uc0/rr2_dev/axi547/ground_gravity/point_datasets/194701.nc'
plot_survey_points(nc_path, 'Freeair', 'gist_heat')
| 0.396185 | 0.402715 |
# Parte 3 do PP1 de RNA2020.1
## Tipos de Tarefas
Recapitulem que dados fornecem experiência sobre um problema. No caso em questão, sugira:
- [x] Uma tarefa de classificação mediante Aprendizado Supervisionado que poderia ser feita com
esta base de dados. Qual seria o atributo-alvo? Quais métricas de desempenho poderiam ser
aplicadas? Que tipo de validação seria apropriado?
- [x] Uma tarefa de regressão mediante Aprendizado Supervisionado que poderia ser feita com
esta base de dados. Qual seria o atributo-alvo? Quais atributos preditores a equipe considera
relevantes para o cenário?
- [x] Bônus: Qual tarefa de Aprendizado Não-Supervisionado poderia ser concebida neste contexto?
```
import warnings
warnings.filterwarnings('ignore')
import re
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import dates
from matplotlib.pyplot import plot_date
%matplotlib inline
plt.style.use('ggplot')
file = 'dataset_limpo_covid19_manaus.csv'
df_dataset = pd.read_csv(file)
df_dataset.dt_notificacao = pd.to_datetime(df_dataset.dt_notificacao)
df_dataset
```
## Visualizando alguns valores dos atributos
```
df_dataset.sexo.value_counts()
# Pie chart, where the slices will be ordered and plotted counter-clockwise:
labels = 'F', 'M'
sizes = df_dataset.sexo.value_counts()
explode = (0, 0.1, 0, 0) # only "explode" the 2nd slice (i.e. 'Hogs')
fig1, ax1 = plt.subplots()
ax1.pie(sizes, labels=labels, autopct='%1.1f%%',
shadow=True, startangle=90)
ax1.axis('equal') # Equal aspect ratio ensures that pie is drawn as a circle.
plt.show()
df_dataset.conclusao.value_counts()
# Pie chart, where the slices will be ordered and plotted counter-clockwise:
labels = 'Recuperados', 'Óbitos'
sizes = df_dataset.conclusao.value_counts()
explode = (0, 0.1, 0, 0) # only "explode" the 2nd slice (i.e. 'Hogs')
fig1, ax1 = plt.subplots()
ax1.pie(sizes, labels=labels, autopct='%1.1f%%',
shadow=True, startangle=90)
ax1.axis('equal') # Equal aspect ratio ensures that pie is drawn as a circle.
plt.show()
df_dataset.idade.value_counts()
x = df_dataset.idade.values
plt.hist(x, density=False, bins=100) # `density=False` would make counts
plt.ylabel('quantidade de pessoas')
plt.xlabel('idade');
# Distribuição das idades por data de confirmação
d = dates.date2num(df_dataset.dt_notificacao)
plot_date(d, df_dataset.idade)
```
## Propondo terefa de ***classificação*** para a base de dados
### Tarefa: classificar se o "paciente" se recuperou ou não do covid-19
Com base nos dados solicitados pelo projeto, uma tarefa de classificação que pode ser realizada em cima dos dados é: classificar um "paciente" como *Recuperado* da covid-19 ou evoluiu para *Óbito*. Um dos problemas encontrados para realizar esta tarefa é o desbalenceamento de clases, pois enquanto a quantidade de "pacientes" recuperados é 6347 o número de pessoas com conclusão igual a Óbito chega a 13.
Uma possível forma de se contornar esse problema do desbalenceamento das clases seria, selecionar outros atributos, pois observa-se que há bastante pessoas classificadas como Óbito, mas por causa da limpeza e escolha desses atributos do dataset atual essa classe diminuiu considerávelmente.
Outra sugestão para aumentar a quantidade de classes minoritária seria considerar mais registros com campos faltantes e depois adicionar a média do atributo para este campo ou outra encontrar outra medida para preencher tais campos.
### Avaliação da tarefa de classificação
Como se trata de uma classificação binária a acurácia é uma das principais métricas a ser considerada, porém para este conjunto de dados talvez não reflita a total realidade por ele apresentar quantidades de classes desbalanceadas, então a melhor maneira é adicionar outras métricas de válidação como por exemplo a precisão que diz quantos acertos para uma classe (true positives) sobre o total de classificados como positivos (true positives + false positives).
Aliados a estas métricas citadas a cima, acredita-se que as seguintes também tornariam a avaliação mais eficiente para o conjuto de dados considerado: *recall*, *f1-score* e também a *G-score* (bastante usada em casses desbalanceadas).
### Validação do modelo
Para a validação do modelo, ou modelos caso se use mais de um para esta tarefa, pensou-se em criar dois experimentos um usando a partição holdout e um outro usando o válidação cruzada. O primeiro para aferir se com os daos é possível criar um bom classificador mesmo que tenha classes desbalanceadas e o segundo para garantir que o/os modelo/os possa ser treinado/os e testado/os com todos os registros e assim contornar o problema das classes desbalanceadas.
## Propondo uma tarefa de ***regressão*** para os dados
### Tarefa: prever a idade dos "pacientes" com base nos demais atributos
Uma das tarefas de regressão pensada para este problema foi a previsão da idade dos pacientes, pois como se base a taxa de letalidade é maior nas pessoas idosas, mas será que com a base de dados disponível usando atributos como bairro, e sexo situação de conclusão do paciente e data de notificação e tipo de test se consegue acertar a idade dele? Nesta tarefa, acreditasse que os atributos tipo_teste e df_notificação não tenham tanha relevância, pois eles não são características dos pacientes, mas sim dos testes que eles fizeram.
## Bônus: Qual tarefa de Aprendizado Não-Supervisionado poderia ser concebida neste contexto?
Considerando o dataset limpo e preprocessado e com quantidade de atributos disponíveis. Uma tarefa de apredizado não supervisionado que poderia ser feita é o agrupamento de perfis de pacientes mais parecidos, ou seja, agrupar os pacientes com base nas suas caracteríscas. Uma meneira de realizar essa tarefa seria o usando o algoritmo *KNN* para realizar esse agrupamento e descobrir os perfis de quem se recuperou ou morreu por exemplo.
|
github_jupyter
|
import warnings
warnings.filterwarnings('ignore')
import re
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import dates
from matplotlib.pyplot import plot_date
%matplotlib inline
plt.style.use('ggplot')
file = 'dataset_limpo_covid19_manaus.csv'
df_dataset = pd.read_csv(file)
df_dataset.dt_notificacao = pd.to_datetime(df_dataset.dt_notificacao)
df_dataset
df_dataset.sexo.value_counts()
# Pie chart, where the slices will be ordered and plotted counter-clockwise:
labels = 'F', 'M'
sizes = df_dataset.sexo.value_counts()
explode = (0, 0.1, 0, 0) # only "explode" the 2nd slice (i.e. 'Hogs')
fig1, ax1 = plt.subplots()
ax1.pie(sizes, labels=labels, autopct='%1.1f%%',
shadow=True, startangle=90)
ax1.axis('equal') # Equal aspect ratio ensures that pie is drawn as a circle.
plt.show()
df_dataset.conclusao.value_counts()
# Pie chart, where the slices will be ordered and plotted counter-clockwise:
labels = 'Recuperados', 'Óbitos'
sizes = df_dataset.conclusao.value_counts()
explode = (0, 0.1, 0, 0) # only "explode" the 2nd slice (i.e. 'Hogs')
fig1, ax1 = plt.subplots()
ax1.pie(sizes, labels=labels, autopct='%1.1f%%',
shadow=True, startangle=90)
ax1.axis('equal') # Equal aspect ratio ensures that pie is drawn as a circle.
plt.show()
df_dataset.idade.value_counts()
x = df_dataset.idade.values
plt.hist(x, density=False, bins=100) # `density=False` would make counts
plt.ylabel('quantidade de pessoas')
plt.xlabel('idade');
# Distribuição das idades por data de confirmação
d = dates.date2num(df_dataset.dt_notificacao)
plot_date(d, df_dataset.idade)
| 0.568775 | 0.924824 |
# `Resource` Wrapper Tutorial
Interactions with MLDB occurs via a [REST API](/doc#builtin/WorkingWithRest.md.html). Interacting with a REST API over HTTP from a Notebook interface can be a little bit laborious if you're using a general-purpose Python library like [`requests`](http://docs.python-requests.org/en/latest/) directly, so MLDB comes with a Python library called [`pymldb`](https://github.com/datacratic/pymldb) to ease the pain.
`pymldb` does this in three ways:
* **the Python `Resource` class**: this is simple class which wraps the `requests` library so as to make HTTP calls to the MLDB API more friendly in a Notebook environment. This tutorial shows you how to use it.
* **the `%mldb` magics**: these are Jupyter line- and cell-magic commands which allow you to make raw HTTP calls to MLDB, and also provides some higher-level functions. Check out the [Cell magic Tutorial](/doc/nblink.html#Cell Magic Tutorial) for more info on the `%mldb` magic system.
* **the Python `BatFrame` class**: this is a class that behaves like the Pandas DataFrame but offloads computation to the server via HTTP calls. Check out the [BatFrame Tutorial](/doc/nblink.html#BatFrame Tutorial) for more info on the BatFrame.
## Getting started
A `Resource` object is just an extremely cheap-to-create, **immutable** proxy for a single URL.
```
from pymldb.resource import Resource
r = Resource("http://localhost")
print r
```
You can use a `Resource` object to quickly create new `Resource` objects to refer to different URLs by calling functions or passing in arguments, chaining the calls:
```
print type(r), r
x = r.x
print type(x), x
y = x("y")
print type(y), y
z = r("and").so("on")("and").so.on
print type(z), z
```
## Making HTTP requests
Once you have a `Resource` object that refers to a URL you care about, you can use it to issue HTTP requests:
```
dataset_types = r.v1.types.datasets
dataset_types.get()
```
The HTTP request is performed via the Python [`requests`](http://docs.python-requests.org/en/latest/) library: arguments to `get()`, `post()`, `put()` and `delete()` are just delegated to the corresponding `requests` function. The only thing that `Resource` does to the result is patch it so it will display prettily in a Notebook, as above.
## Convenience methods
`Resource` objects provide three convenience methods for interacting with MLDB: `get_query()`, `put_json()` and `post_json()`:
```
#keyword arguments to get_query() are appended to the GET query string
r.v1.types.get_query(x="y")
sample_dataset = r.v1.datasets("sample")
sample_dataset.delete()
#dictionaries arguments to put_json() and post_json() are sent as JSON via PUT or POST
sample_dataset.put_json( {"type": "beh.mutable"} )
```
## Putting it all together
Now that you've seen the basics, check out the [Predicting Titanic Survival](/doc/nblink.html#Predicting Titanic Survival) demo to see how to use the `Resource` class to do machine learning with MLDB.
|
github_jupyter
|
from pymldb.resource import Resource
r = Resource("http://localhost")
print r
print type(r), r
x = r.x
print type(x), x
y = x("y")
print type(y), y
z = r("and").so("on")("and").so.on
print type(z), z
dataset_types = r.v1.types.datasets
dataset_types.get()
#keyword arguments to get_query() are appended to the GET query string
r.v1.types.get_query(x="y")
sample_dataset = r.v1.datasets("sample")
sample_dataset.delete()
#dictionaries arguments to put_json() and post_json() are sent as JSON via PUT or POST
sample_dataset.put_json( {"type": "beh.mutable"} )
| 0.270095 | 0.929568 |
# Introduction
Our ultimate aim is to predict solar electricity power generation over the next few hours.
## Loading terabytes of data efficiently from cloud storage
We have several TB of satellite data. To keep the GPU fed with data during training, we need to read chunks of data quickly from the Zarr store; and we also want to load data asynchronously. That is, while the GPU is training on the current batch, the data loader should simultaneously load the _next_ batch from disk.
PyTorch makes this easy! PyTorch's `DataLoader` spawns multiple worker processes when constructed with `num_workers` set to more than 1. Each worker process receives a copy of the `SatelliteDataset` object.
There is a small challenge: The code hangs when it gets to `enumerate(dataloader)` if we open the `xarray.DataArray` in the main process and copy that opened `DataArray` to the child processes. Our solution is to delay the creation of the `DataArray` until _after_ the worker processes have been created. PyTorch makes this easy by allowing us to pass a `worker_init_fn` to `DataLoader`. `worker_init_fn` is called on each worker process. Our `worker_init_fn` just has one job: to call `SatelliteDataset.per_worker_init()` which, in turn, opens the `DataArray`.
This approach achieves read speeds of 600 MB/s from Google Cloud Storage to a single GCP VM with 12 vCPUs (as measured by `nethogs`).
We use `IterableDataset` instead of `Dataset` so `SatelliteDataset` can pre-load the next example from disk and then block (on the `yield`) waiting for PyTorch to read that data. This allows the worker processes to be processing the next data samples while the main process is training the current batch on the GPU.
We can't pin the memory in each worker process because pinned memory can't be shared across processes. Instead we ask `DataLoader` to pin the collated batch so that pytorch-lightning can asynchronously load the next batch from pinned CPU memory into GPU memory.
The satellite data is stored on disk as `int16`. To speed up the movement of the satellite data between processes and from the CPU memory to the GPU memory, we keep the data as `int16` until the data gets to the GPU, where it is converted to `float32` and normalised.
### Loading data from disk into memory in chunks
Cloud storage buckets can't seek into files like 'proper' POSIX filesystems can. So, even if we just want 1 byte from a 1 GB file, we have to load the entire 1 GB file from the bucket.
Zarr is designed with this challenge in mind. Zarr gets round the inability of cloud storage buckets to seek by chunking the data into lots of small files. But, still, we have to load entire Zarr chunks at once, even if we only want a part of a chunk. And, even though we can pull 600 MB/s from a cloud storage bucket, the reads from the storage bucket are still the rate-limiting-step. (GPUs are very fast and have a voracious appetite for data!)
To get the most out of each disk read, our worker processes load several contiguous chunks of Zarr data from disk into memory at once. We then randomly sample from the in-memory data multiple times, before loading another set of chunks from disk into memory. This trick increases training speed by about 10x.
Each Zarr chunk is 36 timesteps long and contains the entire geographical extent. Each timestep is about 5 minutes apart, so each Zarr chunk spans 1.5 hours, assuming the timesteps are contiguous (more on contiguous chunks later).
### Loading only daylight data
We're interested in forecasting solar power generation, so we don't care about nighttime data :)
In the UK in summer, the sun rises first in the north east, and sets last in the north west (see [video of June 2019](https://www.youtube.com/watch?v=IOp-tj-IJpk&t=0s)). In summer, the north gets more hours of sunshine per day.
In the UK in winter, the sun rises first in the south east, and sets last in the south west (see [video of Jan 2019](https://www.youtube.com/watch?v=CJ4prUVa2nQ)). In winter, the south gets more hours of sunshine per day.
| | Summer | Winter |
| ---: | :---: | :---: |
| Sun rises first in | N.E. | S.E. |
| Sun sets last in | N.W. | S.W. |
| Most hours of sunlight | North | South |
We always load a pre-defined number of Zarr chunks from disk every disk load (defined by `n_chunks_per_disk_load`).
Before training, we select timesteps which have at least some sunlight. We do this by computing the clearsky global horizontal irradiance (GHI) for the four corners of the satellite imagery, and for all the timesteps in the dataset. We only use timesteps where the maximum global horizontal irradiance across all four corners is above some threshold.
(The 'clearsky [solar irradiance](https://en.wikipedia.org/wiki/Solar_irradiance)' is the amount of sunlight we'd expect on a clear day at a specific time and location. The SI unit of irradiance is watt per square meter. The 'global horizontal irradiance' is the total sunlight that would hit a horizontal surface on the surface of the Earth. The GHI is the sum of the direct irradiance (sunlight which takes a direct path from the Sun to the Earth's surface) and the diffuse horizontal irradiance (the sunlight scattered from the atmosphere)).
### Finding contiguous sequences
Once we have a list of 'lit' timesteps, we then find contiguous sequences (timeseries without any gaps). And we then compute a list of contiguous Zarr chunks that we'll load at once during training.
### Loading data during training
During training, each worker process randomly picks multiple contiguous Zarr chunk sequences from the list of contiguous sequences pre-computed before training started. The worker loads that data into memory and then randomly samples many samples from that in-memory data before loading more data from disk.
#### Ensuring each batch contains a random sample of the dataset
When PyTorch's `DataLoader` constructs a batch, it reads from just one worker process. (This is not how I had _assumed_ it would work: I assumed PyTorch would construct each batch by randomly sampling from all workers.) This is an issue because, for stochastic gradient descent to work correctly, each batch must contain random samples of the dataset. So it's not sufficient for each worker to load just one contiguous Zarr chunk (because then each batch would be made up entirely of samples from roughly the same time of day). So, instead, each worker process loads multiple contiguous Zarr sequences into memory. This also means that each worker must load quite a lot of data from disk. To avoid training pausing while a worker process loads more data from disk, the data loading is done asynchronously using a separate thread within each worker process.
## Timestep numbering:
* t<sub>0</sub> is 'now'; the most recent observation.
* t<sub>1</sub> is the first timestep of the forecast.
```
# Python core
from typing import Optional, Callable, TypedDict, Union, Iterable, Tuple, NamedTuple, List
from dataclasses import dataclass
import datetime
from itertools import product
from concurrent import futures
# Scientific python
import numpy as np
import pandas as pd
import xarray as xr
import numcodecs
import matplotlib.pyplot as plt
# Cloud compute
import gcsfs
# PyTorch
import torch
from torch import nn
import torch.nn.functional as F
from torchvision import transforms
import pytorch_lightning as pl
# PV & geospatial
import pvlib
import pyproj
```
## Consts & config
The [Zarr docs](https://zarr.readthedocs.io/en/stable/tutorial.html#configuring-blosc) say we should tell the Blosc compression library not to use threads because we're using multiple processes to read from our Zarr store:
```
numcodecs.blosc.use_threads = False
ZARR = 'solar-pv-nowcasting-data/satellite/EUMETSAT/SEVIRI_RSS/OSGB36/all_zarr_int16'
plt.rcParams['figure.figsize'] = (10, 10)
plt.rcParams['image.interpolation'] = 'none'
torch.cuda.is_available()
```
# Load satellite data
```
def get_sat_data(filename: str=ZARR) -> xr.DataArray:
"""Lazily opens the Zarr store on Google Cloud Storage (GCS).
Selects the High Resolution Visible (HRV) satellite channel.
"""
gcs = gcsfs.GCSFileSystem()
store = gcsfs.GCSMap(root=filename, gcs=gcs)
dataset = xr.open_zarr(store, consolidated=True)
return dataset['stacked_eumetsat_data'].sel(variable='HRV')
%%time
sat_data = get_sat_data()
```
Caution: Wierdly, plotting `sat_data` at this point causes the code to hang (with no errors messages) when it gets to `enumerate(dataloader)`. The code hangs even if we first do `sat_data.close(); del sat_data`
```
sat_data
```
Get the timestep indicies of each Zarr chunk. Later, we will use these boundaries to ensure we load complete chunks at a time.
```
zarr_chunk_boundaries = np.concatenate(([0], np.cumsum(sat_data.chunks[0])))
```
## Select daylight hours
```
# OSGB is also called "OSGB 1936 / British National Grid -- United Kingdom Ordnance Survey".
# OSGB is used in many UK electricity system maps, and is used by the UK Met Office UKV model.
# OSGB is a Transverse Mercator projection, using 'easting' and 'northing' coordinates
# which are in meters.
OSGB = 27700
# WGS84 is short for "World Geodetic System 1984", used in GPS. Uses latitude and longitude.
WGS84 = 4326
# osgb_to_wgs84.transform() returns latitude (north-south), longitude (east-west)
osgb_to_wgs84 = pyproj.Transformer.from_crs(crs_from=OSGB, crs_to=WGS84)
def get_daylight_timestamps(
dt_index: pd.DatetimeIndex,
locations: Iterable[Tuple[float, float]],
ghi_threshold: float = 1
) -> pd.DatetimeIndex:
"""Returns datetimes for which the global horizontal irradiance
(GHI) is above ghi_threshold across all locations.
Args:
dt_index: DatetimeIndex to filter. Must be UTC.
locations: List of Tuples of x, y coordinates in OSGB projection.
ghi_threshold: Global horizontal irradiance threshold.
"""
assert dt_index.tz.zone == 'UTC'
ghi_for_all_locations = []
for x, y in locations:
lat, lon = osgb_to_wgs84.transform(x, y)
location = pvlib.location.Location(latitude=lat, longitude=lon)
clearsky = location.get_clearsky(dt_index)
ghi = clearsky['ghi']
ghi_for_all_locations.append(ghi)
ghi_for_all_locations = pd.concat(ghi_for_all_locations, axis='columns')
max_ghi = ghi_for_all_locations.max(axis='columns')
mask = max_ghi > ghi_threshold
return dt_index[mask]
GEO_BORDER: int = 64 #: In same geo projection and units as sat_data.
corners = [
(sat_data.x.values[x], sat_data.y.values[y])
for x, y in product(
[GEO_BORDER, -GEO_BORDER],
[GEO_BORDER, -GEO_BORDER])]
%%time
datetimes = get_daylight_timestamps(
dt_index=pd.DatetimeIndex(sat_data.time.values, tz='UTC'),
locations=corners)
```
## Get contiguous segments of satellite data
```
class Segment(NamedTuple):
"""Represents the start and end indicies of a segment of contiguous samples."""
start: int
end: int
def get_contiguous_segments(dt_index: pd.DatetimeIndex, min_timesteps: int, max_gap: pd.Timedelta) -> Iterable[Segment]:
"""Chunk datetime index into contiguous segments, each at least min_timesteps long.
max_gap defines the threshold for what constitutes a 'gap' between contiguous segments.
Throw away any timesteps in a sequence shorter than min_timesteps long.
"""
gap_mask = np.diff(dt_index) > max_gap
gap_indices = np.argwhere(gap_mask)[:, 0]
# gap_indicies are the indices into dt_index for the timestep immediately before the gap.
# e.g. if the datetimes at 12:00, 12:05, 18:00, 18:05 then gap_indicies will be [1].
segment_boundaries = gap_indices + 1
# Capture the last segment of dt_index.
segment_boundaries = np.concatenate((segment_boundaries, [len(dt_index)]))
segments = []
start_i = 0
for end_i in segment_boundaries:
n_timesteps = end_i - start_i
if n_timesteps >= min_timesteps:
segment = Segment(start=start_i, end=end_i)
segments.append(segment)
start_i = end_i
return segments
%%time
contiguous_segments = get_contiguous_segments(
dt_index = datetimes,
min_timesteps = 36 * 1.5,
max_gap = pd.Timedelta('5 minutes'))
contiguous_segments[:5]
len(contiguous_segments)
```
## Turn the contiguous segments into sequences of Zarr chunks, which will be loaded together during training
```
def get_zarr_chunk_sequences(
n_chunks_per_disk_load: int,
zarr_chunk_boundaries: Iterable[int],
contiguous_segments: Iterable[Segment]) -> Iterable[Segment]:
"""
Args:
n_chunks_per_disk_load: Maximum number of Zarr chunks to load from disk in one go.
zarr_chunk_boundaries: The indicies into the Zarr store's time dimension which define the Zarr chunk boundaries.
Must be sorted.
contiguous_segments: Indicies into the Zarr store's time dimension that define contiguous timeseries.
That is, timeseries with no gaps.
Returns zarr_chunk_sequences: a list of Segments representing the start and end indicies of contiguous sequences of multiple Zarr chunks,
all exactly n_chunks_per_disk_load long (for contiguous segments at least as long as n_chunks_per_disk_load zarr chunks),
and at least one side of the boundary will lie on a 'natural' Zarr chunk boundary.
For example, say that n_chunks_per_disk_load = 3, and the Zarr chunks sizes are all 5:
0 5 10 15 20 25 30 35
|....|....|....|....|....|....|....|
INPUTS:
|------CONTIGUOUS SEGMENT----|
zarr_chunk_boundaries:
|----|----|----|----|----|----|----|
OUTPUT:
zarr_chunk_sequences:
3 to 15: |-|----|----|
5 to 20: |----|----|----|
10 to 25: |----|----|----|
15 to 30: |----|----|----|
20 to 32: |----|----|-|
"""
assert n_chunks_per_disk_load > 0
zarr_chunk_sequences = []
for contig_segment in contiguous_segments:
# searchsorted() returns the index into zarr_chunk_boundaries at which contig_segment.start
# should be inserted into zarr_chunk_boundaries to maintain a sorted list.
# i_of_first_zarr_chunk is the index to the element in zarr_chunk_boundaries which defines
# the start of the current contig chunk.
i_of_first_zarr_chunk = np.searchsorted(zarr_chunk_boundaries, contig_segment.start)
# i_of_first_zarr_chunk will be too large by 1 unless contig_segment.start lies
# exactly on a Zarr chunk boundary. Hence we must subtract 1, or else we'll
# end up with the first contig_chunk being 1 + n_chunks_per_disk_load chunks long.
if zarr_chunk_boundaries[i_of_first_zarr_chunk] > contig_segment.start:
i_of_first_zarr_chunk -= 1
# Prepare for looping to create multiple Zarr chunk sequences for the current contig_segment.
zarr_chunk_seq_start_i = contig_segment.start
zarr_chunk_seq_end_i = None # Just a convenience to allow us to break the while loop by checking if zarr_chunk_seq_end_i != contig_segment.end.
while zarr_chunk_seq_end_i != contig_segment.end:
zarr_chunk_seq_end_i = zarr_chunk_boundaries[i_of_first_zarr_chunk + n_chunks_per_disk_load]
zarr_chunk_seq_end_i = min(zarr_chunk_seq_end_i, contig_segment.end)
zarr_chunk_sequences.append(Segment(start=zarr_chunk_seq_start_i, end=zarr_chunk_seq_end_i))
i_of_first_zarr_chunk += 1
zarr_chunk_seq_start_i = zarr_chunk_boundaries[i_of_first_zarr_chunk]
return zarr_chunk_sequences
zarr_chunk_sequences = get_zarr_chunk_sequences(
n_chunks_per_disk_load=3,
zarr_chunk_boundaries=zarr_chunk_boundaries,
contiguous_segments=contiguous_segments)
zarr_chunk_sequences[:10]
```
## PyTorch data storage & processing
```
Array = Union[np.ndarray, xr.DataArray]
IMAGE_ATTR_NAMES = ('historical_sat_images', 'target_sat_images')
class Sample(TypedDict):
"""Simple class for structuring data for the ML model.
Using typing.TypedDict gives us several advantages:
1. Single 'source of truth' for the type and documentation of each example.
2. A static type checker can check the types are correct.
Instead of TypedDict, we could use typing.NamedTuple,
which would provide runtime checks, but the deal-breaker with Tuples is that they're immutable
so we cannot change the values in the transforms.
"""
# IMAGES
# Shape: batch_size, seq_length, width, height
historical_sat_images: Array
target_sat_images: Array
# METADATA
datetime_index: Array
class BadData(Exception):
pass
@dataclass
class RandomSquareCrop():
size: int = 128 #: Size of the cropped image.
def __call__(self, sample: Sample) -> Sample:
crop_params = None
for attr_name in IMAGE_ATTR_NAMES:
image = sample[attr_name]
# TODO: Random crop!
cropped_image = image[..., :self.size, :self.size]
sample[attr_name] = cropped_image
return sample
class CheckForBadData():
def __call__(self, sample: Sample) -> Sample:
for attr_name in IMAGE_ATTR_NAMES:
image = sample[attr_name]
if np.any(image < 0):
raise BadData(f'\n{attr_name} has negative values at {image.time.values}')
return sample
class ToTensor():
def __call__(self, sample: Sample) -> Sample:
for key, value in sample.items():
if isinstance(value, xr.DataArray):
value = value.values
sample[key] = torch.from_numpy(value)
return sample
```
## PyTorch dataset
```
@dataclass
class SatelliteDataset(torch.utils.data.IterableDataset):
zarr_chunk_sequences: Iterable[Segment] #: Defines multiple Zarr chunks to be loaded from disk at once.
history_len: int = 1 #: The number of timesteps of 'history' to load.
forecast_len: int = 1 #: The number of timesteps of 'forecast' to load.
transform: Optional[Callable] = None
n_disk_loads_per_epoch: int = 10_000 #: Number of disk loads per worker process per epoch.
min_n_samples_per_disk_load: int = 1_000 #: Number of samples each worker will load for each disk load.
max_n_samples_per_disk_load: int = 2_000 #: Max number of disk loader. Actual number is chosen randomly between min & max.
n_zarr_chunk_sequences_to_load_at_once: int = 8 #: Number of chunk seqs to load at once. These are sampled at random.
def __post_init__(self):
#: Total sequence length of each sample.
self.total_seq_len = self.history_len + self.forecast_len
def per_worker_init(self, worker_id: int) -> None:
"""Called by worker_init_fn on each copy of SatelliteDataset after the worker process has been spawned."""
self.worker_id = worker_id
self.data_array = get_sat_data()
# Each worker must have a different seed for its random number generator.
# Otherwise all the workers will output exactly the same data!
seed = torch.initial_seed()
self.rng = np.random.default_rng(seed=seed)
def __iter__(self):
"""
Asynchronously loads next data from disk while sampling from data_in_mem.
"""
with futures.ThreadPoolExecutor(max_workers=1) as executor:
future_data = executor.submit(self._load_data_from_disk)
for _ in range(self.n_disk_loads_per_epoch):
data_in_mem = future_data.result()
future_data = executor.submit(self._load_data_from_disk)
n_samples = self.rng.integers(self.min_n_samples_per_disk_load, self.max_n_samples_per_disk_load)
for _ in range(n_samples):
sample = self._get_sample(data_in_mem)
if self.transform:
try:
sample = self.transform(sample)
except BadData as e:
print(e)
continue
yield sample
def _load_data_from_disk(self) -> List[xr.DataArray]:
"""Loads data from contiguous Zarr chunks from disk into memory."""
sat_images_list = []
for _ in range(self.n_zarr_chunk_sequences_to_load_at_once):
zarr_chunk_sequence = self.rng.choice(self.zarr_chunk_sequences)
sat_images = self.data_array.isel(time=slice(*zarr_chunk_sequence))
# Sanity checks
n_timesteps_available = len(sat_images)
if n_timesteps_available < self.total_seq_len:
raise RuntimeError(f'Not enough timesteps in loaded data! Need at least {self.total_seq_len}. Got {n_timesteps_available}!')
sat_images_list.append(sat_images.load())
return sat_images_list
def _get_sample(self, data_in_mem_list: List[xr.DataArray]) -> Sample:
i = self.rng.integers(0, len(data_in_mem_list))
data_in_mem = data_in_mem_list[i]
n_timesteps_available = len(data_in_mem)
max_start_idx = n_timesteps_available - self.total_seq_len
start_idx = self.rng.integers(low=0, high=max_start_idx, dtype=np.uint32)
end_idx = start_idx + self.total_seq_len
sat_images = data_in_mem.isel(time=slice(start_idx, end_idx))
return Sample(
historical_sat_images=sat_images[:self.history_len],
target_sat_images=sat_images[self.history_len:],
datetime_index=sat_images.time.values.astype('datetime64[s]').astype(int)
)
def worker_init_fn(worker_id):
"""Configures each dataset worker process.
Just has one job! To call SatelliteDataset.per_worker_init().
"""
# get_worker_info() returns information specific to each worker process.
worker_info = torch.utils.data.get_worker_info()
if worker_info is None:
print('worker_info is None!')
else:
dataset_obj = worker_info.dataset # The Dataset copy in this worker process.
dataset_obj.per_worker_init(worker_id=worker_info.id)
torch.manual_seed(42)
dataset = SatelliteDataset(
zarr_chunk_sequences=zarr_chunk_sequences,
transform=transforms.Compose([
RandomSquareCrop(),
CheckForBadData(),
ToTensor(),
]),
)
dataloader = torch.utils.data.DataLoader(
dataset,
batch_size=8,
num_workers=8, # timings: 4=13.8s; 8=11.6; 10=11.3s; 11=11.5s; 12=12.6s. 10=3it/s
worker_init_fn=worker_init_fn,
pin_memory=True,
#persistent_workers=True
)
%%time
for i, batch in enumerate(dataloader):
print(i, batch['historical_sat_images'].shape)
break
pd.to_datetime(batch['datetime_index'].numpy().flatten(), unit='s').values.reshape(-1, 2).astype('datetime64[s]')
batch['historical_sat_images'].shape
batch['target_sat_images'].shape
batch['historical_sat_images'].dtype
plt.imshow(batch['historical_sat_images'][4, 0])
```
# Simple ML model
```
def normalise_images_in_model(images, device):
SAT_IMAGE_MEAN = torch.tensor(93.23458, dtype=torch.float, device=device)
SAT_IMAGE_STD = torch.tensor(115.34247, dtype=torch.float, device=device)
images = images.float()
images -= SAT_IMAGE_MEAN
images /= SAT_IMAGE_STD
return images
CHANNELS = 32
KERNEL = 3
class LitAutoEncoder(pl.LightningModule):
def __init__(self):
super().__init__()
self.encoder_conv1 = nn.Conv2d(in_channels=1, out_channels=CHANNELS//2, kernel_size=KERNEL)
self.encoder_conv2 = nn.Conv2d(in_channels=CHANNELS//2, out_channels=CHANNELS, kernel_size=KERNEL)
self.encoder_conv3 = nn.Conv2d(in_channels=CHANNELS, out_channels=CHANNELS, kernel_size=KERNEL)
self.encoder_conv4 = nn.Conv2d(in_channels=CHANNELS, out_channels=CHANNELS, kernel_size=KERNEL)
self.maxpool = nn.MaxPool2d(kernel_size=KERNEL)
self.decoder_conv1 = nn.ConvTranspose2d(in_channels=CHANNELS, out_channels=CHANNELS, kernel_size=KERNEL)
self.decoder_conv2 = nn.ConvTranspose2d(in_channels=CHANNELS, out_channels=CHANNELS//2, kernel_size=KERNEL)
self.decoder_conv3 = nn.ConvTranspose2d(in_channels=CHANNELS//2, out_channels=CHANNELS//2, kernel_size=KERNEL)
self.decoder_conv4 = nn.ConvTranspose2d(in_channels=CHANNELS//2, out_channels=1, kernel_size=KERNEL)
def forward(self, x):
images = x['historical_sat_images']
images = normalise_images_in_model(images, self.device)
# Pass data through the network :)
# ENCODER
out = F.relu(self.encoder_conv1(images))
out = F.relu(self.encoder_conv2(out))
out = F.relu(self.encoder_conv3(out))
out = F.relu(self.encoder_conv4(out))
out = self.maxpool(out)
# DECODER
out = F.relu(self.decoder_conv1(out))
out = F.relu(self.decoder_conv2(out))
out = F.relu(self.decoder_conv3(out))
out = self.decoder_conv4(out)
return out
def _training_or_validation_step(self, batch, is_train_step):
y_hat = self(batch)
y = batch['target_sat_images']
y = normalise_images_in_model(y, self.device)
y = y[..., 40:-40, 40:-40] # Due to the CNN stride, the output image is 48 x 48
loss = F.mse_loss(y_hat, y)
tag = "Loss/Train" if is_train_step else "Loss/Validation"
self.log_dict({tag: loss}, on_step=is_train_step, on_epoch=True)
return loss
def training_step(self, batch, batch_idx):
return self._training_or_validation_step(batch, is_train_step=True)
def validation_step(self, batch, batch_idx):
return self._training_or_validation_step(batch, is_train_step=False)
def configure_optimizers(self):
optimizer = torch.optim.Adam(self.parameters(), lr=0.001)
return optimizer
model = LitAutoEncoder()
trainer = pl.Trainer(gpus=1, max_epochs=400, terminate_on_nan=False)
%%time
trainer.fit(model, train_dataloader=dataloader)
```
|
github_jupyter
|
# Python core
from typing import Optional, Callable, TypedDict, Union, Iterable, Tuple, NamedTuple, List
from dataclasses import dataclass
import datetime
from itertools import product
from concurrent import futures
# Scientific python
import numpy as np
import pandas as pd
import xarray as xr
import numcodecs
import matplotlib.pyplot as plt
# Cloud compute
import gcsfs
# PyTorch
import torch
from torch import nn
import torch.nn.functional as F
from torchvision import transforms
import pytorch_lightning as pl
# PV & geospatial
import pvlib
import pyproj
numcodecs.blosc.use_threads = False
ZARR = 'solar-pv-nowcasting-data/satellite/EUMETSAT/SEVIRI_RSS/OSGB36/all_zarr_int16'
plt.rcParams['figure.figsize'] = (10, 10)
plt.rcParams['image.interpolation'] = 'none'
torch.cuda.is_available()
def get_sat_data(filename: str=ZARR) -> xr.DataArray:
"""Lazily opens the Zarr store on Google Cloud Storage (GCS).
Selects the High Resolution Visible (HRV) satellite channel.
"""
gcs = gcsfs.GCSFileSystem()
store = gcsfs.GCSMap(root=filename, gcs=gcs)
dataset = xr.open_zarr(store, consolidated=True)
return dataset['stacked_eumetsat_data'].sel(variable='HRV')
%%time
sat_data = get_sat_data()
sat_data
zarr_chunk_boundaries = np.concatenate(([0], np.cumsum(sat_data.chunks[0])))
# OSGB is also called "OSGB 1936 / British National Grid -- United Kingdom Ordnance Survey".
# OSGB is used in many UK electricity system maps, and is used by the UK Met Office UKV model.
# OSGB is a Transverse Mercator projection, using 'easting' and 'northing' coordinates
# which are in meters.
OSGB = 27700
# WGS84 is short for "World Geodetic System 1984", used in GPS. Uses latitude and longitude.
WGS84 = 4326
# osgb_to_wgs84.transform() returns latitude (north-south), longitude (east-west)
osgb_to_wgs84 = pyproj.Transformer.from_crs(crs_from=OSGB, crs_to=WGS84)
def get_daylight_timestamps(
dt_index: pd.DatetimeIndex,
locations: Iterable[Tuple[float, float]],
ghi_threshold: float = 1
) -> pd.DatetimeIndex:
"""Returns datetimes for which the global horizontal irradiance
(GHI) is above ghi_threshold across all locations.
Args:
dt_index: DatetimeIndex to filter. Must be UTC.
locations: List of Tuples of x, y coordinates in OSGB projection.
ghi_threshold: Global horizontal irradiance threshold.
"""
assert dt_index.tz.zone == 'UTC'
ghi_for_all_locations = []
for x, y in locations:
lat, lon = osgb_to_wgs84.transform(x, y)
location = pvlib.location.Location(latitude=lat, longitude=lon)
clearsky = location.get_clearsky(dt_index)
ghi = clearsky['ghi']
ghi_for_all_locations.append(ghi)
ghi_for_all_locations = pd.concat(ghi_for_all_locations, axis='columns')
max_ghi = ghi_for_all_locations.max(axis='columns')
mask = max_ghi > ghi_threshold
return dt_index[mask]
GEO_BORDER: int = 64 #: In same geo projection and units as sat_data.
corners = [
(sat_data.x.values[x], sat_data.y.values[y])
for x, y in product(
[GEO_BORDER, -GEO_BORDER],
[GEO_BORDER, -GEO_BORDER])]
%%time
datetimes = get_daylight_timestamps(
dt_index=pd.DatetimeIndex(sat_data.time.values, tz='UTC'),
locations=corners)
class Segment(NamedTuple):
"""Represents the start and end indicies of a segment of contiguous samples."""
start: int
end: int
def get_contiguous_segments(dt_index: pd.DatetimeIndex, min_timesteps: int, max_gap: pd.Timedelta) -> Iterable[Segment]:
"""Chunk datetime index into contiguous segments, each at least min_timesteps long.
max_gap defines the threshold for what constitutes a 'gap' between contiguous segments.
Throw away any timesteps in a sequence shorter than min_timesteps long.
"""
gap_mask = np.diff(dt_index) > max_gap
gap_indices = np.argwhere(gap_mask)[:, 0]
# gap_indicies are the indices into dt_index for the timestep immediately before the gap.
# e.g. if the datetimes at 12:00, 12:05, 18:00, 18:05 then gap_indicies will be [1].
segment_boundaries = gap_indices + 1
# Capture the last segment of dt_index.
segment_boundaries = np.concatenate((segment_boundaries, [len(dt_index)]))
segments = []
start_i = 0
for end_i in segment_boundaries:
n_timesteps = end_i - start_i
if n_timesteps >= min_timesteps:
segment = Segment(start=start_i, end=end_i)
segments.append(segment)
start_i = end_i
return segments
%%time
contiguous_segments = get_contiguous_segments(
dt_index = datetimes,
min_timesteps = 36 * 1.5,
max_gap = pd.Timedelta('5 minutes'))
contiguous_segments[:5]
len(contiguous_segments)
def get_zarr_chunk_sequences(
n_chunks_per_disk_load: int,
zarr_chunk_boundaries: Iterable[int],
contiguous_segments: Iterable[Segment]) -> Iterable[Segment]:
"""
Args:
n_chunks_per_disk_load: Maximum number of Zarr chunks to load from disk in one go.
zarr_chunk_boundaries: The indicies into the Zarr store's time dimension which define the Zarr chunk boundaries.
Must be sorted.
contiguous_segments: Indicies into the Zarr store's time dimension that define contiguous timeseries.
That is, timeseries with no gaps.
Returns zarr_chunk_sequences: a list of Segments representing the start and end indicies of contiguous sequences of multiple Zarr chunks,
all exactly n_chunks_per_disk_load long (for contiguous segments at least as long as n_chunks_per_disk_load zarr chunks),
and at least one side of the boundary will lie on a 'natural' Zarr chunk boundary.
For example, say that n_chunks_per_disk_load = 3, and the Zarr chunks sizes are all 5:
0 5 10 15 20 25 30 35
|....|....|....|....|....|....|....|
INPUTS:
|------CONTIGUOUS SEGMENT----|
zarr_chunk_boundaries:
|----|----|----|----|----|----|----|
OUTPUT:
zarr_chunk_sequences:
3 to 15: |-|----|----|
5 to 20: |----|----|----|
10 to 25: |----|----|----|
15 to 30: |----|----|----|
20 to 32: |----|----|-|
"""
assert n_chunks_per_disk_load > 0
zarr_chunk_sequences = []
for contig_segment in contiguous_segments:
# searchsorted() returns the index into zarr_chunk_boundaries at which contig_segment.start
# should be inserted into zarr_chunk_boundaries to maintain a sorted list.
# i_of_first_zarr_chunk is the index to the element in zarr_chunk_boundaries which defines
# the start of the current contig chunk.
i_of_first_zarr_chunk = np.searchsorted(zarr_chunk_boundaries, contig_segment.start)
# i_of_first_zarr_chunk will be too large by 1 unless contig_segment.start lies
# exactly on a Zarr chunk boundary. Hence we must subtract 1, or else we'll
# end up with the first contig_chunk being 1 + n_chunks_per_disk_load chunks long.
if zarr_chunk_boundaries[i_of_first_zarr_chunk] > contig_segment.start:
i_of_first_zarr_chunk -= 1
# Prepare for looping to create multiple Zarr chunk sequences for the current contig_segment.
zarr_chunk_seq_start_i = contig_segment.start
zarr_chunk_seq_end_i = None # Just a convenience to allow us to break the while loop by checking if zarr_chunk_seq_end_i != contig_segment.end.
while zarr_chunk_seq_end_i != contig_segment.end:
zarr_chunk_seq_end_i = zarr_chunk_boundaries[i_of_first_zarr_chunk + n_chunks_per_disk_load]
zarr_chunk_seq_end_i = min(zarr_chunk_seq_end_i, contig_segment.end)
zarr_chunk_sequences.append(Segment(start=zarr_chunk_seq_start_i, end=zarr_chunk_seq_end_i))
i_of_first_zarr_chunk += 1
zarr_chunk_seq_start_i = zarr_chunk_boundaries[i_of_first_zarr_chunk]
return zarr_chunk_sequences
zarr_chunk_sequences = get_zarr_chunk_sequences(
n_chunks_per_disk_load=3,
zarr_chunk_boundaries=zarr_chunk_boundaries,
contiguous_segments=contiguous_segments)
zarr_chunk_sequences[:10]
Array = Union[np.ndarray, xr.DataArray]
IMAGE_ATTR_NAMES = ('historical_sat_images', 'target_sat_images')
class Sample(TypedDict):
"""Simple class for structuring data for the ML model.
Using typing.TypedDict gives us several advantages:
1. Single 'source of truth' for the type and documentation of each example.
2. A static type checker can check the types are correct.
Instead of TypedDict, we could use typing.NamedTuple,
which would provide runtime checks, but the deal-breaker with Tuples is that they're immutable
so we cannot change the values in the transforms.
"""
# IMAGES
# Shape: batch_size, seq_length, width, height
historical_sat_images: Array
target_sat_images: Array
# METADATA
datetime_index: Array
class BadData(Exception):
pass
@dataclass
class RandomSquareCrop():
size: int = 128 #: Size of the cropped image.
def __call__(self, sample: Sample) -> Sample:
crop_params = None
for attr_name in IMAGE_ATTR_NAMES:
image = sample[attr_name]
# TODO: Random crop!
cropped_image = image[..., :self.size, :self.size]
sample[attr_name] = cropped_image
return sample
class CheckForBadData():
def __call__(self, sample: Sample) -> Sample:
for attr_name in IMAGE_ATTR_NAMES:
image = sample[attr_name]
if np.any(image < 0):
raise BadData(f'\n{attr_name} has negative values at {image.time.values}')
return sample
class ToTensor():
def __call__(self, sample: Sample) -> Sample:
for key, value in sample.items():
if isinstance(value, xr.DataArray):
value = value.values
sample[key] = torch.from_numpy(value)
return sample
@dataclass
class SatelliteDataset(torch.utils.data.IterableDataset):
zarr_chunk_sequences: Iterable[Segment] #: Defines multiple Zarr chunks to be loaded from disk at once.
history_len: int = 1 #: The number of timesteps of 'history' to load.
forecast_len: int = 1 #: The number of timesteps of 'forecast' to load.
transform: Optional[Callable] = None
n_disk_loads_per_epoch: int = 10_000 #: Number of disk loads per worker process per epoch.
min_n_samples_per_disk_load: int = 1_000 #: Number of samples each worker will load for each disk load.
max_n_samples_per_disk_load: int = 2_000 #: Max number of disk loader. Actual number is chosen randomly between min & max.
n_zarr_chunk_sequences_to_load_at_once: int = 8 #: Number of chunk seqs to load at once. These are sampled at random.
def __post_init__(self):
#: Total sequence length of each sample.
self.total_seq_len = self.history_len + self.forecast_len
def per_worker_init(self, worker_id: int) -> None:
"""Called by worker_init_fn on each copy of SatelliteDataset after the worker process has been spawned."""
self.worker_id = worker_id
self.data_array = get_sat_data()
# Each worker must have a different seed for its random number generator.
# Otherwise all the workers will output exactly the same data!
seed = torch.initial_seed()
self.rng = np.random.default_rng(seed=seed)
def __iter__(self):
"""
Asynchronously loads next data from disk while sampling from data_in_mem.
"""
with futures.ThreadPoolExecutor(max_workers=1) as executor:
future_data = executor.submit(self._load_data_from_disk)
for _ in range(self.n_disk_loads_per_epoch):
data_in_mem = future_data.result()
future_data = executor.submit(self._load_data_from_disk)
n_samples = self.rng.integers(self.min_n_samples_per_disk_load, self.max_n_samples_per_disk_load)
for _ in range(n_samples):
sample = self._get_sample(data_in_mem)
if self.transform:
try:
sample = self.transform(sample)
except BadData as e:
print(e)
continue
yield sample
def _load_data_from_disk(self) -> List[xr.DataArray]:
"""Loads data from contiguous Zarr chunks from disk into memory."""
sat_images_list = []
for _ in range(self.n_zarr_chunk_sequences_to_load_at_once):
zarr_chunk_sequence = self.rng.choice(self.zarr_chunk_sequences)
sat_images = self.data_array.isel(time=slice(*zarr_chunk_sequence))
# Sanity checks
n_timesteps_available = len(sat_images)
if n_timesteps_available < self.total_seq_len:
raise RuntimeError(f'Not enough timesteps in loaded data! Need at least {self.total_seq_len}. Got {n_timesteps_available}!')
sat_images_list.append(sat_images.load())
return sat_images_list
def _get_sample(self, data_in_mem_list: List[xr.DataArray]) -> Sample:
i = self.rng.integers(0, len(data_in_mem_list))
data_in_mem = data_in_mem_list[i]
n_timesteps_available = len(data_in_mem)
max_start_idx = n_timesteps_available - self.total_seq_len
start_idx = self.rng.integers(low=0, high=max_start_idx, dtype=np.uint32)
end_idx = start_idx + self.total_seq_len
sat_images = data_in_mem.isel(time=slice(start_idx, end_idx))
return Sample(
historical_sat_images=sat_images[:self.history_len],
target_sat_images=sat_images[self.history_len:],
datetime_index=sat_images.time.values.astype('datetime64[s]').astype(int)
)
def worker_init_fn(worker_id):
"""Configures each dataset worker process.
Just has one job! To call SatelliteDataset.per_worker_init().
"""
# get_worker_info() returns information specific to each worker process.
worker_info = torch.utils.data.get_worker_info()
if worker_info is None:
print('worker_info is None!')
else:
dataset_obj = worker_info.dataset # The Dataset copy in this worker process.
dataset_obj.per_worker_init(worker_id=worker_info.id)
torch.manual_seed(42)
dataset = SatelliteDataset(
zarr_chunk_sequences=zarr_chunk_sequences,
transform=transforms.Compose([
RandomSquareCrop(),
CheckForBadData(),
ToTensor(),
]),
)
dataloader = torch.utils.data.DataLoader(
dataset,
batch_size=8,
num_workers=8, # timings: 4=13.8s; 8=11.6; 10=11.3s; 11=11.5s; 12=12.6s. 10=3it/s
worker_init_fn=worker_init_fn,
pin_memory=True,
#persistent_workers=True
)
%%time
for i, batch in enumerate(dataloader):
print(i, batch['historical_sat_images'].shape)
break
pd.to_datetime(batch['datetime_index'].numpy().flatten(), unit='s').values.reshape(-1, 2).astype('datetime64[s]')
batch['historical_sat_images'].shape
batch['target_sat_images'].shape
batch['historical_sat_images'].dtype
plt.imshow(batch['historical_sat_images'][4, 0])
def normalise_images_in_model(images, device):
SAT_IMAGE_MEAN = torch.tensor(93.23458, dtype=torch.float, device=device)
SAT_IMAGE_STD = torch.tensor(115.34247, dtype=torch.float, device=device)
images = images.float()
images -= SAT_IMAGE_MEAN
images /= SAT_IMAGE_STD
return images
CHANNELS = 32
KERNEL = 3
class LitAutoEncoder(pl.LightningModule):
def __init__(self):
super().__init__()
self.encoder_conv1 = nn.Conv2d(in_channels=1, out_channels=CHANNELS//2, kernel_size=KERNEL)
self.encoder_conv2 = nn.Conv2d(in_channels=CHANNELS//2, out_channels=CHANNELS, kernel_size=KERNEL)
self.encoder_conv3 = nn.Conv2d(in_channels=CHANNELS, out_channels=CHANNELS, kernel_size=KERNEL)
self.encoder_conv4 = nn.Conv2d(in_channels=CHANNELS, out_channels=CHANNELS, kernel_size=KERNEL)
self.maxpool = nn.MaxPool2d(kernel_size=KERNEL)
self.decoder_conv1 = nn.ConvTranspose2d(in_channels=CHANNELS, out_channels=CHANNELS, kernel_size=KERNEL)
self.decoder_conv2 = nn.ConvTranspose2d(in_channels=CHANNELS, out_channels=CHANNELS//2, kernel_size=KERNEL)
self.decoder_conv3 = nn.ConvTranspose2d(in_channels=CHANNELS//2, out_channels=CHANNELS//2, kernel_size=KERNEL)
self.decoder_conv4 = nn.ConvTranspose2d(in_channels=CHANNELS//2, out_channels=1, kernel_size=KERNEL)
def forward(self, x):
images = x['historical_sat_images']
images = normalise_images_in_model(images, self.device)
# Pass data through the network :)
# ENCODER
out = F.relu(self.encoder_conv1(images))
out = F.relu(self.encoder_conv2(out))
out = F.relu(self.encoder_conv3(out))
out = F.relu(self.encoder_conv4(out))
out = self.maxpool(out)
# DECODER
out = F.relu(self.decoder_conv1(out))
out = F.relu(self.decoder_conv2(out))
out = F.relu(self.decoder_conv3(out))
out = self.decoder_conv4(out)
return out
def _training_or_validation_step(self, batch, is_train_step):
y_hat = self(batch)
y = batch['target_sat_images']
y = normalise_images_in_model(y, self.device)
y = y[..., 40:-40, 40:-40] # Due to the CNN stride, the output image is 48 x 48
loss = F.mse_loss(y_hat, y)
tag = "Loss/Train" if is_train_step else "Loss/Validation"
self.log_dict({tag: loss}, on_step=is_train_step, on_epoch=True)
return loss
def training_step(self, batch, batch_idx):
return self._training_or_validation_step(batch, is_train_step=True)
def validation_step(self, batch, batch_idx):
return self._training_or_validation_step(batch, is_train_step=False)
def configure_optimizers(self):
optimizer = torch.optim.Adam(self.parameters(), lr=0.001)
return optimizer
model = LitAutoEncoder()
trainer = pl.Trainer(gpus=1, max_epochs=400, terminate_on_nan=False)
%%time
trainer.fit(model, train_dataloader=dataloader)
| 0.911308 | 0.983629 |
```
import warnings
warnings.filterwarnings('ignore')
import numpy as np
import pandas as pd
from pathlib import Path
from collections import Counter
from sklearn.metrics import balanced_accuracy_score
from sklearn.metrics import confusion_matrix
from imblearn.metrics import classification_report_imbalanced
```
# Read the CSV and Perform Basic Data Cleaning
```
# https://help.lendingclub.com/hc/en-us/articles/215488038-What-do-the-different-Note-statuses-mean-
columns = [
"loan_amnt", "int_rate", "installment", "home_ownership",
"annual_inc", "verification_status", "issue_d", "loan_status",
"pymnt_plan", "dti", "delinq_2yrs", "inq_last_6mths",
"open_acc", "pub_rec", "revol_bal", "total_acc",
"initial_list_status", "out_prncp", "out_prncp_inv", "total_pymnt",
"total_pymnt_inv", "total_rec_prncp", "total_rec_int", "total_rec_late_fee",
"recoveries", "collection_recovery_fee", "last_pymnt_amnt", "next_pymnt_d",
"collections_12_mths_ex_med", "policy_code", "application_type", "acc_now_delinq",
"tot_coll_amt", "tot_cur_bal", "open_acc_6m", "open_act_il",
"open_il_12m", "open_il_24m", "mths_since_rcnt_il", "total_bal_il",
"il_util", "open_rv_12m", "open_rv_24m", "max_bal_bc",
"all_util", "total_rev_hi_lim", "inq_fi", "total_cu_tl",
"inq_last_12m", "acc_open_past_24mths", "avg_cur_bal", "bc_open_to_buy",
"bc_util", "chargeoff_within_12_mths", "delinq_amnt", "mo_sin_old_il_acct",
"mo_sin_old_rev_tl_op", "mo_sin_rcnt_rev_tl_op", "mo_sin_rcnt_tl", "mort_acc",
"mths_since_recent_bc", "mths_since_recent_inq", "num_accts_ever_120_pd", "num_actv_bc_tl",
"num_actv_rev_tl", "num_bc_sats", "num_bc_tl", "num_il_tl",
"num_op_rev_tl", "num_rev_accts", "num_rev_tl_bal_gt_0",
"num_sats", "num_tl_120dpd_2m", "num_tl_30dpd", "num_tl_90g_dpd_24m",
"num_tl_op_past_12m", "pct_tl_nvr_dlq", "percent_bc_gt_75", "pub_rec_bankruptcies",
"tax_liens", "tot_hi_cred_lim", "total_bal_ex_mort", "total_bc_limit",
"total_il_high_credit_limit", "hardship_flag", "debt_settlement_flag"
]
target = ["loan_status"]
# Load the data
file_path = Path('Resources/LoanStats_2019Q1.csv')
df = pd.read_csv(file_path, skiprows=1)[:-2]
df = df.loc[:, columns].copy()
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
# Remove the `Issued` loan status
issued_mask = df['loan_status'] != 'Issued'
df = df.loc[issued_mask]
# convert interest rate to numerical
df['int_rate'] = df['int_rate'].str.replace('%', '')
df['int_rate'] = df['int_rate'].astype('float') / 100
# Convert the target column values to low_risk and high_risk based on their values
x = {'Current': 'low_risk'}
df = df.replace(x)
x = dict.fromkeys(['Late (31-120 days)', 'Late (16-30 days)', 'Default', 'In Grace Period'], 'high_risk')
df = df.replace(x)
df.reset_index(inplace=True, drop=True)
df.head()
```
# Split the Data into Training and Testing
```
# Create our features
X = pd.get_dummies(df.drop('loan_status', axis=1))
# Create our target
y = df['loan_status'].tolist()
X.describe()
# Check the balance of our target values
df['loan_status'].value_counts()
# Split the X and y into X_train, X_test, y_train, y_test
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test= train_test_split(X,
y,
random_state=1,
stratify=y)
X_train.shape
```
## Data Pre-Processing
Scale the training and testing data using the `StandardScaler` from `sklearn`. Remember that when scaling the data, you only scale the features data (`X_train` and `X_testing`).
```
# Create the StandardScaler instance
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
# Fit the Standard Scaler with the training data
# When fitting scaling functions, only train on the training dataset
X_scaler = scaler.fit(X_train)
# Scale the training and testing data
X_train_scaled = X_scaler.transform(X_train)
X_test_scaled = X_scaler.transform(X_test)
```
# Ensemble Learners
In this section, you will compare two ensemble algorithms to determine which algorithm results in the best performance. You will train a Balanced Random Forest Classifier and an Easy Ensemble classifier . For each algorithm, be sure to complete the folliowing steps:
1. Train the model using the training data.
2. Calculate the balanced accuracy score from sklearn.metrics.
3. Print the confusion matrix from sklearn.metrics.
4. Generate a classication report using the `imbalanced_classification_report` from imbalanced-learn.
5. For the Balanced Random Forest Classifier onely, print the feature importance sorted in descending order (most important feature to least important) along with the feature score
Note: Use a random state of 1 for each algorithm to ensure consistency between tests
### Balanced Random Forest Classifier
```
# Resample the training data with the BalancedRandomForestClassifier
from imblearn.ensemble import BalancedRandomForestClassifier
rf=BalancedRandomForestClassifier(random_state=1,class_weight='balanced',sampling_strategy='not minority')
rf.fit(X_train, y_train)
# Calculated the balanced accuracy score
predictions = rf.predict(X_test)
balanced_accuracy_score(y_test, predictions)
# Display the confusion matrix
predictions = rf.predict(X_test)
results = pd.DataFrame({"Prediction": predictions,
"Actual": y_test}).reset_index(drop=True)
results.head()
confusion_matrix(y_test, predictions)
# Print the imbalanced classification report
y_pred_rf = rf.predict(X_test)
print(classification_report_imbalanced(y_test, y_pred_rf))
# List the features sorted in descending order by feature importance
feature_importances = pd.DataFrame(rf.feature_importances_,
index = X_train.columns,
columns=['importance']).sort_values('importance', ascending=False)
print(feature_importances)
```
### Easy Ensemble Classifier
```
#Train the Classifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn import datasets
adaboost = AdaBoostClassifier(n_estimators=1000,
learning_rate=1,random_state=1)
model = adaboost.fit(X_train, y_train)
y_pred = model.predict(X_test)
# Calculated the balanced accuracy score
from sklearn.metrics import balanced_accuracy_score
balanced_accuracy_score(y_test, y_pred)
# Display the confusion matrix
confusion_matrix(y_test, y_pred)
# Print the imbalanced classification report
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
```
|
github_jupyter
|
import warnings
warnings.filterwarnings('ignore')
import numpy as np
import pandas as pd
from pathlib import Path
from collections import Counter
from sklearn.metrics import balanced_accuracy_score
from sklearn.metrics import confusion_matrix
from imblearn.metrics import classification_report_imbalanced
# https://help.lendingclub.com/hc/en-us/articles/215488038-What-do-the-different-Note-statuses-mean-
columns = [
"loan_amnt", "int_rate", "installment", "home_ownership",
"annual_inc", "verification_status", "issue_d", "loan_status",
"pymnt_plan", "dti", "delinq_2yrs", "inq_last_6mths",
"open_acc", "pub_rec", "revol_bal", "total_acc",
"initial_list_status", "out_prncp", "out_prncp_inv", "total_pymnt",
"total_pymnt_inv", "total_rec_prncp", "total_rec_int", "total_rec_late_fee",
"recoveries", "collection_recovery_fee", "last_pymnt_amnt", "next_pymnt_d",
"collections_12_mths_ex_med", "policy_code", "application_type", "acc_now_delinq",
"tot_coll_amt", "tot_cur_bal", "open_acc_6m", "open_act_il",
"open_il_12m", "open_il_24m", "mths_since_rcnt_il", "total_bal_il",
"il_util", "open_rv_12m", "open_rv_24m", "max_bal_bc",
"all_util", "total_rev_hi_lim", "inq_fi", "total_cu_tl",
"inq_last_12m", "acc_open_past_24mths", "avg_cur_bal", "bc_open_to_buy",
"bc_util", "chargeoff_within_12_mths", "delinq_amnt", "mo_sin_old_il_acct",
"mo_sin_old_rev_tl_op", "mo_sin_rcnt_rev_tl_op", "mo_sin_rcnt_tl", "mort_acc",
"mths_since_recent_bc", "mths_since_recent_inq", "num_accts_ever_120_pd", "num_actv_bc_tl",
"num_actv_rev_tl", "num_bc_sats", "num_bc_tl", "num_il_tl",
"num_op_rev_tl", "num_rev_accts", "num_rev_tl_bal_gt_0",
"num_sats", "num_tl_120dpd_2m", "num_tl_30dpd", "num_tl_90g_dpd_24m",
"num_tl_op_past_12m", "pct_tl_nvr_dlq", "percent_bc_gt_75", "pub_rec_bankruptcies",
"tax_liens", "tot_hi_cred_lim", "total_bal_ex_mort", "total_bc_limit",
"total_il_high_credit_limit", "hardship_flag", "debt_settlement_flag"
]
target = ["loan_status"]
# Load the data
file_path = Path('Resources/LoanStats_2019Q1.csv')
df = pd.read_csv(file_path, skiprows=1)[:-2]
df = df.loc[:, columns].copy()
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
# Remove the `Issued` loan status
issued_mask = df['loan_status'] != 'Issued'
df = df.loc[issued_mask]
# convert interest rate to numerical
df['int_rate'] = df['int_rate'].str.replace('%', '')
df['int_rate'] = df['int_rate'].astype('float') / 100
# Convert the target column values to low_risk and high_risk based on their values
x = {'Current': 'low_risk'}
df = df.replace(x)
x = dict.fromkeys(['Late (31-120 days)', 'Late (16-30 days)', 'Default', 'In Grace Period'], 'high_risk')
df = df.replace(x)
df.reset_index(inplace=True, drop=True)
df.head()
# Create our features
X = pd.get_dummies(df.drop('loan_status', axis=1))
# Create our target
y = df['loan_status'].tolist()
X.describe()
# Check the balance of our target values
df['loan_status'].value_counts()
# Split the X and y into X_train, X_test, y_train, y_test
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test= train_test_split(X,
y,
random_state=1,
stratify=y)
X_train.shape
# Create the StandardScaler instance
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
# Fit the Standard Scaler with the training data
# When fitting scaling functions, only train on the training dataset
X_scaler = scaler.fit(X_train)
# Scale the training and testing data
X_train_scaled = X_scaler.transform(X_train)
X_test_scaled = X_scaler.transform(X_test)
# Resample the training data with the BalancedRandomForestClassifier
from imblearn.ensemble import BalancedRandomForestClassifier
rf=BalancedRandomForestClassifier(random_state=1,class_weight='balanced',sampling_strategy='not minority')
rf.fit(X_train, y_train)
# Calculated the balanced accuracy score
predictions = rf.predict(X_test)
balanced_accuracy_score(y_test, predictions)
# Display the confusion matrix
predictions = rf.predict(X_test)
results = pd.DataFrame({"Prediction": predictions,
"Actual": y_test}).reset_index(drop=True)
results.head()
confusion_matrix(y_test, predictions)
# Print the imbalanced classification report
y_pred_rf = rf.predict(X_test)
print(classification_report_imbalanced(y_test, y_pred_rf))
# List the features sorted in descending order by feature importance
feature_importances = pd.DataFrame(rf.feature_importances_,
index = X_train.columns,
columns=['importance']).sort_values('importance', ascending=False)
print(feature_importances)
#Train the Classifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn import datasets
adaboost = AdaBoostClassifier(n_estimators=1000,
learning_rate=1,random_state=1)
model = adaboost.fit(X_train, y_train)
y_pred = model.predict(X_test)
# Calculated the balanced accuracy score
from sklearn.metrics import balanced_accuracy_score
balanced_accuracy_score(y_test, y_pred)
# Display the confusion matrix
confusion_matrix(y_test, y_pred)
# Print the imbalanced classification report
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
| 0.615435 | 0.738551 |
# Data Exploration
Learning objectives:
1. Learn useful patterns for exploring data before modeling
2. Gain an understanding of the dataset and identify any data issues.
The goal of this notebook is to explore our base tables before we began feature engineering and modeling. We will explore the price history of stock in the S&P 500.
* Price history : Price history of stocks
* S&P 500 : A list of all companies and symbols for companies in the S&P 500
For our analysis, let's limit price history since 2000. In general, the further back historical data is used the lower it's predictive power can be.
```
import os
PROJECT = !(gcloud config get-value core/project)
PROJECT = PROJECT[0]
BUCKET = PROJECT
os.environ['PROJECT'] = PROJECT
os.environ['BUCKET'] = BUCKET
import numpy as np
import pandas as pd
import seaborn as sns
from matplotlib import pyplot as plt
from google.cloud import bigquery
from IPython.core.magic import register_cell_magic
from IPython import get_ipython
bq = bigquery.Client(project=PROJECT)
# Allow you to easily have Python variables in SQL query.
@register_cell_magic('with_globals')
def with_globals(line, cell):
contents = cell.format(**globals())
if 'print' in line:
print(contents)
get_ipython().run_cell(contents)
```
## Preparing the dataset
Let's create the dataset in our project BiqQuery and import the stock data by running the following cells:
```
!bq mk stock_src
%%bash
TABLE=price_history
SCHEMA=symbol:STRING,Date:DATE,Open:FLOAT,Close:FLOAT
test -f $TABLE.csv || unzip ../stock_src/$TABLE.csv.zip
gsutil -m cp $TABLE.csv gs://$BUCKET/stock_src/$TABLE.csv
bq load --source_format=CSV --skip_leading_rows=1 \
stock_src.$TABLE gs://$BUCKET/stock_src/$TABLE.csv $SCHEMA
%%bash
TABLE=eps
SCHEMA=date:DATE,company:STRING,symbol:STRING,surprise:STRING,reported_EPS:FLOAT,consensus_EPS:FLOAT
test -f $TABLE.csv || unzip ../stock_src/$TABLE.csv.zip
gsutil -m cp $TABLE.csv gs://$BUCKET/stock_src/$TABLE.csv
bq load --source_format=CSV --skip_leading_rows=1 \
stock_src.$TABLE gs://$BUCKET/stock_src/$TABLE.csv $SCHEMA
%%bash
TABLE=snp500
SCHEMA=company:STRING,symbol:STRING,industry:STRING
test -f $TABLE.csv || unzip ../stock_src/$TABLE.csv.zip
gsutil -m cp $TABLE.csv gs://$BUCKET/stock_src/$TABLE.csv
bq load --source_format=CSV --skip_leading_rows=1 \
stock_src.$TABLE gs://$BUCKET/stock_src/$TABLE.csv $SCHEMA
```
Let's look at the tables and columns we have for analysis.
**Learning objective 1.**
```
%%with_globals
%%bigquery --project {PROJECT}
SELECT table_name, column_name, data_type
FROM `stock_src.INFORMATION_SCHEMA.COLUMNS`
ORDER BY table_name, ordinal_position
```
## Price History
Retrieve Google's stock price history.
```
def query_stock(symbol):
return bq.query('''
SELECT *
FROM `stock_src.price_history`
WHERE symbol="{0}"
ORDER BY Date
'''.format(symbol)).to_dataframe()
df_stock = query_stock('GOOG')
df_stock.Date = pd.to_datetime(df_stock.Date)
ax = df_stock.plot(x='Date', y='Close', title='Google stock')
# Add smoothed plot.
df_stock['Close_smoothed'] = df_stock.Close.rolling(100, center=True).mean()
df_stock.plot(x='Date', y='Close_smoothed', ax=ax);
```
Compare google to S&P
```
df_sp = query_stock('gspc')
def plot_with_sp(symbol):
df_stock = query_stock(symbol)
df_stock.Date = pd.to_datetime(df_stock.Date)
df_stock.Date = pd.to_datetime(df_stock.Date)
fig = plt.figure()
ax1 = fig.add_subplot(111)
ax2 = ax1.twinx()
ax = df_sp.plot(x='Date', y='Close', label='S&P', color='green', ax=ax1,
alpha=0.7)
ax = df_stock.plot(x='Date', y='Close', label=symbol,
title=symbol + ' and S&P index', ax=ax2, alpha=0.7)
ax1.legend(loc=3)
ax2.legend(loc=4)
ax1.set_ylabel('S&P price')
ax2.set_ylabel(symbol + ' price')
ax.set_xlim(pd.to_datetime('2004-08-05'), pd.to_datetime('2013-08-05'))
plot_with_sp('GOOG')
```
**Learning objective 2**
```
plot_with_sp('IBM')
```
Let's see how the price of stocks change over time on a yearly basis. Using the `LAG` function we can compute the change in stock price year-over-year.
Let's compute average close difference for each year. This line could, of course, be done in Pandas. Often times it's useful to use some combination of BigQuery and Pandas for exploration analysis. In general, it's most effective to let BigQuery do the heavy-duty processing and then use Pandas for smaller data and visualization.
**Learning objective 1, 2**
```
%%with_globals
%%bigquery df --project {PROJECT}
WITH
with_year AS
(
SELECT symbol,
EXTRACT(YEAR FROM date) AS year,
close
FROM `stock_src.price_history`
WHERE symbol in (SELECT symbol FROM `stock_src.snp500`)
),
year_aggregated AS
(
SELECT year, symbol, AVG(close) as avg_close
FROM with_year
WHERE year >= 2000
GROUP BY year, symbol
)
SELECT year, symbol, avg_close as close,
(LAG(avg_close, 1) OVER (PARTITION BY symbol order by year DESC))
AS next_yr_close
FROM year_aggregated
ORDER BY symbol, year
```
Compute the year-over-year percentage increase.
```
df.dropna(inplace=True)
df['percent_increase'] = (df.next_yr_close - df.close) / df.close
```
Let's visualize some yearly stock
```
def get_random_stocks(n=5):
random_stocks = df.symbol.sample(n=n, random_state=3)
rand = df.merge(random_stocks)
return rand[['year', 'symbol', 'percent_increase']]
rand = get_random_stocks()
for symbol, _df in rand.groupby('symbol'):
plt.figure()
sns.barplot(x='year', y="percent_increase", data=_df)
plt.title(symbol)
```
There have been some major fluctations in individual stocks. For example, there were major drops during the early 2000's for tech companies.
```
df.sort_values('percent_increase').head()
stock_symbol = 'YHOO'
%%with_globals
%%bigquery df --project {PROJECT}
SELECT date, close
FROM `stock_src.price_history`
WHERE symbol='{stock_symbol}'
ORDER BY date
ax = df.plot(x='date', y='close')
```
**Stock splits** can also impact our data - causing a stock price to rapidly drop. In practice, we would need to clean all of our stock data to account for this. This would be a major effort! Fortunately, in the case of [IBM](https://www.fool.com/investing/2017/01/06/ibm-stock-split-will-2017-finally-be-the-year-shar.aspx), for example, all stock splits occurred before the year 2000.
**Learning objective 2**
```
stock_symbol = 'IBM'
%%with_globals
%%bigquery df --project {PROJECT}
SELECT date, close
FROM `stock_src.price_history`
WHERE symbol='{stock_symbol}'
ORDER BY date
IBM_STOCK_SPLIT_DATE = '1979-05-10'
ax = df.plot(x='date', y='close')
ax.vlines(pd.to_datetime(IBM_STOCK_SPLIT_DATE),
0, 500, linestyle='dashed', color='grey', alpha=0.7);
```
## S&P companies list
```
%%with_globals
%%bigquery df --project {PROJECT}
SELECT *
FROM `stock_src.snp500`
df.industry.value_counts().plot(kind='barh');
```
We can join the price histories table with the S&P 500 table to compare industries:
**Learning objective 1,2**
```
%%with_globals
%%bigquery df --project {PROJECT}
WITH sp_prices AS
(
SELECT a.*, b.industry
FROM `stock_src.price_history` a
JOIN `stock_src.snp500` b
USING (symbol)
WHERE date >= "2000-01-01"
)
SELECT Date, industry, AVG(close) as close
FROM sp_prices
GROUP BY Date, industry
ORDER BY industry, Date
df.head()
```
Using pandas we can "unstack" our table so that each industry has it's own column. This will be useful for plotting.
```
# Pandas `unstack` to make each industry a column. Useful for plotting.
df_ind = df.set_index(['industry', 'Date']).unstack(0).dropna()
df_ind.columns = [c[1] for c in df_ind.columns]
df_ind.head()
ax = df_ind.plot(figsize=(16, 8))
# Move legend down.
ax.legend(loc='upper center', bbox_to_anchor=(0.5, -0.05), shadow=True, ncol=2)
```
Let's scale each industry using min/max scaling. This will put all of the stocks on the same scale. Currently it can be hard to see the changes in stocks over time across industries.
**Learning objective 1**
```
def min_max_scale(df):
return (df - df.min()) / df.max()
scaled = min_max_scale(df_ind)
ax = scaled.plot(figsize=(16, 8))
ax.legend(loc='upper center', bbox_to_anchor=(0.5, -0.05), shadow=True, ncol=2);
```
We can also create a smoothed version of the plot above using a [rolling mean](https://en.wikipedia.org/wiki/Moving_average). This is a useful transformation to make when visualizing time-series data.
```
SMOOTHING_WINDOW = 30 # Days.
rolling = scaled.copy()
for col in scaled.columns:
rolling[col] = scaled[col].rolling(SMOOTHING_WINDOW).mean()
ax = rolling.plot(figsize=(16, 8))
ax.legend(loc='upper center', bbox_to_anchor=(0.5, -0.05), shadow=True, ncol=2);
```
Information technology had a large crash during the early 2000s and again in 2008/2009; along with all other stocks. After 2008, some industries were a bit slower to recover than other industries.
BONUS: In the next lab, we will want to predict the price of the stock in the future. What are some features that we can use to predict future price? Try visualizing some of these features.
Copyright 2019 Google Inc. Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License
|
github_jupyter
|
import os
PROJECT = !(gcloud config get-value core/project)
PROJECT = PROJECT[0]
BUCKET = PROJECT
os.environ['PROJECT'] = PROJECT
os.environ['BUCKET'] = BUCKET
import numpy as np
import pandas as pd
import seaborn as sns
from matplotlib import pyplot as plt
from google.cloud import bigquery
from IPython.core.magic import register_cell_magic
from IPython import get_ipython
bq = bigquery.Client(project=PROJECT)
# Allow you to easily have Python variables in SQL query.
@register_cell_magic('with_globals')
def with_globals(line, cell):
contents = cell.format(**globals())
if 'print' in line:
print(contents)
get_ipython().run_cell(contents)
!bq mk stock_src
%%bash
TABLE=price_history
SCHEMA=symbol:STRING,Date:DATE,Open:FLOAT,Close:FLOAT
test -f $TABLE.csv || unzip ../stock_src/$TABLE.csv.zip
gsutil -m cp $TABLE.csv gs://$BUCKET/stock_src/$TABLE.csv
bq load --source_format=CSV --skip_leading_rows=1 \
stock_src.$TABLE gs://$BUCKET/stock_src/$TABLE.csv $SCHEMA
%%bash
TABLE=eps
SCHEMA=date:DATE,company:STRING,symbol:STRING,surprise:STRING,reported_EPS:FLOAT,consensus_EPS:FLOAT
test -f $TABLE.csv || unzip ../stock_src/$TABLE.csv.zip
gsutil -m cp $TABLE.csv gs://$BUCKET/stock_src/$TABLE.csv
bq load --source_format=CSV --skip_leading_rows=1 \
stock_src.$TABLE gs://$BUCKET/stock_src/$TABLE.csv $SCHEMA
%%bash
TABLE=snp500
SCHEMA=company:STRING,symbol:STRING,industry:STRING
test -f $TABLE.csv || unzip ../stock_src/$TABLE.csv.zip
gsutil -m cp $TABLE.csv gs://$BUCKET/stock_src/$TABLE.csv
bq load --source_format=CSV --skip_leading_rows=1 \
stock_src.$TABLE gs://$BUCKET/stock_src/$TABLE.csv $SCHEMA
%%with_globals
%%bigquery --project {PROJECT}
SELECT table_name, column_name, data_type
FROM `stock_src.INFORMATION_SCHEMA.COLUMNS`
ORDER BY table_name, ordinal_position
def query_stock(symbol):
return bq.query('''
SELECT *
FROM `stock_src.price_history`
WHERE symbol="{0}"
ORDER BY Date
'''.format(symbol)).to_dataframe()
df_stock = query_stock('GOOG')
df_stock.Date = pd.to_datetime(df_stock.Date)
ax = df_stock.plot(x='Date', y='Close', title='Google stock')
# Add smoothed plot.
df_stock['Close_smoothed'] = df_stock.Close.rolling(100, center=True).mean()
df_stock.plot(x='Date', y='Close_smoothed', ax=ax);
df_sp = query_stock('gspc')
def plot_with_sp(symbol):
df_stock = query_stock(symbol)
df_stock.Date = pd.to_datetime(df_stock.Date)
df_stock.Date = pd.to_datetime(df_stock.Date)
fig = plt.figure()
ax1 = fig.add_subplot(111)
ax2 = ax1.twinx()
ax = df_sp.plot(x='Date', y='Close', label='S&P', color='green', ax=ax1,
alpha=0.7)
ax = df_stock.plot(x='Date', y='Close', label=symbol,
title=symbol + ' and S&P index', ax=ax2, alpha=0.7)
ax1.legend(loc=3)
ax2.legend(loc=4)
ax1.set_ylabel('S&P price')
ax2.set_ylabel(symbol + ' price')
ax.set_xlim(pd.to_datetime('2004-08-05'), pd.to_datetime('2013-08-05'))
plot_with_sp('GOOG')
plot_with_sp('IBM')
%%with_globals
%%bigquery df --project {PROJECT}
WITH
with_year AS
(
SELECT symbol,
EXTRACT(YEAR FROM date) AS year,
close
FROM `stock_src.price_history`
WHERE symbol in (SELECT symbol FROM `stock_src.snp500`)
),
year_aggregated AS
(
SELECT year, symbol, AVG(close) as avg_close
FROM with_year
WHERE year >= 2000
GROUP BY year, symbol
)
SELECT year, symbol, avg_close as close,
(LAG(avg_close, 1) OVER (PARTITION BY symbol order by year DESC))
AS next_yr_close
FROM year_aggregated
ORDER BY symbol, year
df.dropna(inplace=True)
df['percent_increase'] = (df.next_yr_close - df.close) / df.close
def get_random_stocks(n=5):
random_stocks = df.symbol.sample(n=n, random_state=3)
rand = df.merge(random_stocks)
return rand[['year', 'symbol', 'percent_increase']]
rand = get_random_stocks()
for symbol, _df in rand.groupby('symbol'):
plt.figure()
sns.barplot(x='year', y="percent_increase", data=_df)
plt.title(symbol)
df.sort_values('percent_increase').head()
stock_symbol = 'YHOO'
%%with_globals
%%bigquery df --project {PROJECT}
SELECT date, close
FROM `stock_src.price_history`
WHERE symbol='{stock_symbol}'
ORDER BY date
ax = df.plot(x='date', y='close')
stock_symbol = 'IBM'
%%with_globals
%%bigquery df --project {PROJECT}
SELECT date, close
FROM `stock_src.price_history`
WHERE symbol='{stock_symbol}'
ORDER BY date
IBM_STOCK_SPLIT_DATE = '1979-05-10'
ax = df.plot(x='date', y='close')
ax.vlines(pd.to_datetime(IBM_STOCK_SPLIT_DATE),
0, 500, linestyle='dashed', color='grey', alpha=0.7);
%%with_globals
%%bigquery df --project {PROJECT}
SELECT *
FROM `stock_src.snp500`
df.industry.value_counts().plot(kind='barh');
%%with_globals
%%bigquery df --project {PROJECT}
WITH sp_prices AS
(
SELECT a.*, b.industry
FROM `stock_src.price_history` a
JOIN `stock_src.snp500` b
USING (symbol)
WHERE date >= "2000-01-01"
)
SELECT Date, industry, AVG(close) as close
FROM sp_prices
GROUP BY Date, industry
ORDER BY industry, Date
df.head()
# Pandas `unstack` to make each industry a column. Useful for plotting.
df_ind = df.set_index(['industry', 'Date']).unstack(0).dropna()
df_ind.columns = [c[1] for c in df_ind.columns]
df_ind.head()
ax = df_ind.plot(figsize=(16, 8))
# Move legend down.
ax.legend(loc='upper center', bbox_to_anchor=(0.5, -0.05), shadow=True, ncol=2)
def min_max_scale(df):
return (df - df.min()) / df.max()
scaled = min_max_scale(df_ind)
ax = scaled.plot(figsize=(16, 8))
ax.legend(loc='upper center', bbox_to_anchor=(0.5, -0.05), shadow=True, ncol=2);
SMOOTHING_WINDOW = 30 # Days.
rolling = scaled.copy()
for col in scaled.columns:
rolling[col] = scaled[col].rolling(SMOOTHING_WINDOW).mean()
ax = rolling.plot(figsize=(16, 8))
ax.legend(loc='upper center', bbox_to_anchor=(0.5, -0.05), shadow=True, ncol=2);
| 0.431584 | 0.929504 |
# Actividad Limpieza de los Datasets Relacionados con el Indice de Educación.
## Acerca del proyecto:
Este proyecto tiene como proposito realizar un analisis de la calidad de vida de ciertos paises, con el fin de determinar cuál de ellos podria ser la mejor opcion para vivir o buscar un empleo, todo esto, haciendo uso de herramientas estadísticas y computacionales, siendo el principal aliado, el lenguaje de programacion Python.
En este caso se tendrá como principal objetivo calcular el **IDH** por medio de la documentación proporcionada por la **UNDP** en el documento [__*Human Development Report 2020*__](http://hdr.undp.org/sites/default/files/hdr2020_technical_notes.pdf). Y con tales resultados justificar nuestro anterior [proyecto](https://github.com/Team-17-Bedu/proyecto) elaborado en el lenguaje R.
## Notas
Una de las consideraciones realizadas antes de cargar los datasets __*"Expected years of schooling (years).csv"*__ y __*"Mean years of schooling (years).csv"*__. Fue que se hizo una exploración manual a los archivos .csv.
En este analisis se observaron las siguientes peculiaridades en el documento __*"Mean years of schooling (years).csv"*__:
* Contaba con datos extra al principio del documento .csv, como se observa en la siguiente imagen:

<p align="center">
<a>
<img src="https://drive.google.com/uc?export=download&id=1QnFcvVbXZDi83gaH8RupyJNuJSzJuSLU">
</a>
</p>
<br>
Figura 1. Cabecera original
* Contaba con datos extra al final del documento .csv, como se observa en la siguiente imagen:

<p align="center">
<a>
<img src="https://drive.google.com/uc?export=download&id=1rxUjLp0viVp_AwHLzFkYRlXldjGlecLp">
</a>
</p>
<br>
Figura 2. Cola original
Para ello se decidio, suprimir esos datos inecesarios, con el fin de no tener problemas al cargar el .csv en Jupyter Notebook.
## Descripción de la actividad
En esta actividad se pretende hacer un analisis de los datasets __*"Expected years of schooling (years).csv"*__ y __*"Mean years of schooling (years).csv"*__ y una limpieza con el fin de calcular el indice de Educación por país, para posteriormente calcular el IDH.
En un instante se describiran ciertas actividades realizadas al documento .csv:
## Expectativa de años de escolaridad
1. Se realizaron las importaciones necesarias.
```
# Importar modulos necesarios
import pandas as pd
```
2. Lectura del dataset `Expected years of schooling (years).csv` en un `DataFrame` denominado `expectativa`
```
expectativa = pd.read_csv("Expected years of schooling (years).csv", encoding= "latin1")
```
3. Obtención de la información del `DataFrame`
* Visualización del `DataFrame` resultante (tanto `head` como `tail`)
```
expectativa
```
* Dimensiones del `DataFrame`
* 192 filas
* 62 columnas
```
expectativa.shape
```
* Nombre de las columnas
```
expectativa.columns
```
#### Observación
Se tomo la desición de renombrar las columnas, para hacerlas mas comprensibles.
```
head = {'HDI Rank':'Ranking',
'Country':'Pais',
'1990':'1990',
'Unnamed: 3':'Indice_Educacion_1990',
'1991':'1991',
'Unnamed: 5':'Indice_Educacion_1991',
'1992':'1992',
'Unnamed: 7':'Indice_Educacion_1992',
'1993':'1993',
'Unnamed: 9':'Indice_Educacion_1993',
'1994':'1994',
'Unnamed: 11':'Indice_Educacion_1994',
'1995':'1995',
'Unnamed: 13':'Indice_Educacion_1995',
'1996':'1996',
'Unnamed: 15':'Indice_Educacion_1996',
'1997':'1997',
'Unnamed: 17':'Indice_Educacion_1997',
'1998':'1998',
'Unnamed: 19':'Indice_Educacion_1998',
'1999':'1999',
'Unnamed: 21':'Indice_Educacion_1999',
'2000':'2000',
'Unnamed: 23':'Indice_Educacion_2000',
'2001':'2001',
'Unnamed: 25':'Indice_Educacion_2001',
'2002':'2002',
'Unnamed: 27':'Indice_Educacion_2002',
'2003':'2003',
'Unnamed: 29':'Indice_Educacion_2003',
'2004':'2004',
'Unnamed: 31':'Indice_Educacion_2004',
'2005':'2005',
'Unnamed: 33':'Indice_Educacion_2005',
'2006':'2006',
'Unnamed: 35':'Indice_Educacion_2006',
'2007':'2007',
'Unnamed: 37':'Indice_Educacion_2007',
'2008':'2008',
'Unnamed: 39':'Indice_Educacion_2008',
'2009':'2009',
'Unnamed: 41':'Indice_Educacion_2009',
'2010':'2010',
'Unnamed: 43':'Indice_Educacion_2010',
'2011':'2011',
'Unnamed: 45':'Indice_Educacion_2011',
'2012':'2012',
'Unnamed: 47':'Indice_Educacion_2012',
'2013':'2013',
'Unnamed: 49':'Indice_Educacion_2013',
'2014':'2014',
'Unnamed: 51':'Indice_Educacion_2014',
'2015':'2015',
'Unnamed: 53':'Indice_Educacion_2015',
'2016':'2016',
'Unnamed: 55':'Indice_Educacion_2016',
'2017':'2017',
'Unnamed: 57':'Indice_Educacion_2017',
'2018':'2018',
'Unnamed: 59':'Indice_Educacion_2018',
'2019':'2019',
'Unnamed: 61':'Indice_Educacion_2019'}
expectativa = expectativa.rename(columns=head)
```
#### Resultado del cambio de cabeceras
```
expectativa.head()
```
### Calculo del indice de expectativa de años de escolaridad por paises
Se explica a continuación como se realizo el calculo del indice de expectativa de años de escolaaridad por pais.
Primeramente se presenta lo que es la formula que se uso:
$$\Huge\Huge\frac{\alpha - \theta}{\gamma - \theta}$$
Donde:
* $\alpha$ : La expectativa de escolaridad en el pais.
* $\theta$ : Es la expectativa minima de años de escolaridad. En el documento __*Human Development Report 2020*__ se usa 0 años como minimo.
* $\gamma$ : Es la expectativa maxima de años de escolaridad. En el documento __*Human Development Report 2020*__ se usa 18 años como maximo.
Este calculo se realizo por cada pais en cada uno de los años.
```
for year in range(1990, 2020):
year = str(year)
indices = []
for cell in expectativa[year]:
try:
indices.append((float(cell) - 0) / (18 - 0))
except:
indices.append(0)
expectativa[f'Indice_Educacion_{year}'] = indices
expectativa = expectativa.drop([str(i) for i in range(1990, 2020)],axis=1)
```
Resultado del calculo del indice de la expectativa de años de escolaridad por cada país en cada uno de sus periodos.
```
expectativa.head()
```
## Promedio de años de escolaridad
1. Lectura del dataset `Expected years of schooling (years).csv` en un `DataFrame` denominado `expectativa`
```
promedio = pd.read_csv("Mean years of schooling (years).csv")
```
2. Obtención de la información del `DataFrame`
* Visualización del `DataFrame` resultante (tanto `head` como `tail`)
```
promedio
```
* Dimensiones del `DataFrame`
* 190 filas
* 62 columnas
```
promedio.shape
```
* Nombre de las columnas
```
promedio.columns
```
#### Observación
Se tomo la desición de renombrar las columnas, para hacerlas mas comprensibles.
```
head = {'HDI Rank':'Ranking',
'Country':'Pais',
'1990':'1990',
'Unnamed: 3':'Indice_Promedio_Edu_1990',
'1991':'1991',
'Unnamed: 5':'Indice_Promedio_Edu_1991',
'1992':'1992',
'Unnamed: 7':'Indice_Promedio_Edu_1992',
'1993':'1993',
'Unnamed: 9':'Indice_Promedio_Edu_1993',
'1994':'1994',
'Unnamed: 11':'Indice_Promedio_Edu_1994',
'1995':'1995',
'Unnamed: 13':'Indice_Promedio_Edu_1995',
'1996':'1996',
'Unnamed: 15':'Indice_Promedio_Edu_1996',
'1997':'1997',
'Unnamed: 17':'Indice_Promedio_Edu_1997',
'1998':'1998',
'Unnamed: 19':'Indice_Promedio_Edu_1998',
'1999':'1999',
'Unnamed: 21':'Indice_Promedio_Edu_1999',
'2000':'2000',
'Unnamed: 23':'Indice_Promedio_Edu_2000',
'2001':'2001',
'Unnamed: 25':'Indice_Promedio_Edu_2001',
'2002':'2002',
'Unnamed: 27':'Indice_Promedio_Edu_2002',
'2003':'2003',
'Unnamed: 29':'Indice_Promedio_Edu_2003',
'2004':'2004',
'Unnamed: 31':'Indice_Promedio_Edu_2004',
'2005':'2005',
'Unnamed: 33':'Indice_Promedio_Edu_2005',
'2006':'2006',
'Unnamed: 35':'Indice_Promedio_Edu_2006',
'2007':'2007',
'Unnamed: 37':'Indice_Promedio_Edu_2007',
'2008':'2008',
'Unnamed: 39':'Indice_Promedio_Edu_2008',
'2009':'2009',
'Unnamed: 41':'Indice_Promedio_Edu_2009',
'2010':'2010',
'Unnamed: 43':'Indice_Promedio_Edu_2010',
'2011':'2011',
'Unnamed: 45':'Indice_Promedio_Edu_2011',
'2012':'2012',
'Unnamed: 47':'Indice_Promedio_Edu_2012',
'2013':'2013',
'Unnamed: 49':'Indice_Promedio_Edu_2013',
'2014':'2014',
'Unnamed: 51':'Indice_Promedio_Edu_2014',
'2015':'2015',
'Unnamed: 53':'Indice_Promedio_Edu_2015',
'2016':'2016',
'Unnamed: 55':'Indice_Promedio_Edu_2016',
'2017':'2017',
'Unnamed: 57':'Indice_Promedio_Edu_2017',
'2018':'2018',
'Unnamed: 59':'Indice_Promedio_Edu_2018',
'2019':'2019',
'Unnamed: 61':'Indice_Promedio_Edu_2019'}
promedio = promedio.rename(columns=head)
```
#### Resultado del cambio de cabeceras
```
promedio.head()
```
### Calculo del indice de promedio de años de escolaridad por país
Se explica a continuación como se realizo el calculo del indice de promedio de años de escolaridad por pais.
Primeramente se presenta lo que es la formula que se uso:
$$\Huge\Huge\frac{\alpha - \theta}{\gamma - \theta}$$
Donde:
* $\alpha$ : El promedio de años de escolaridad en el pais.
* $\theta$ : Es la expectativa minima del promedio de años de escolaridad. En el documento __*Human Development Report 2020*__ se usa 0 años como minimo.
* $\gamma$ : Es la expectativa maxima del promedio de años de escolaridad. En el documento __*Human Development Report 2020*__ se usa 15 años como maximo.
Este calculo se realizo por cada pais en cada uno de los años.
```
import numpy as np
for year in range(1990, 2020):
year = str(year)
indices = []
for cell in promedio[year]:
try:
indices.append((float(cell) - 0) / (15 - 0))
except:
indices.append(0)
promedio[f'Indice_Promedio_Edu_{year}'] = indices
promedio = promedio.drop([str(i) for i in range(1990, 2020)], axis=1)
```
Resultado del calculo del indice del promedio de años de escolaridad por cada país en cada uno de sus periodos.
```
promedio.head()
```
## Calculo del Indice de Educacion
1. Creación de un `DataFrame` vacío para almacenar todos los resultados del calculo
```
header = ['Pais']
indices = pd.DataFrame({}, columns=header)
```
### Calculo del indice de Educación por país
Se explica a continuación como se realizo el calculo del indice de educación por país.
Primeramente se presenta lo que es la formula que se uso:
$$\Huge\Huge\frac{\alpha + \beta}{2}$$
Donde:
* $\alpha$ : El indice de la expectativa de años de escolaridad en el pais.
* $\beta$ : El indice del promedio de años de escolaridad en el país
Este calculo se realizo por cada pais en cada uno de los años.
```
import numpy as np
def calcular_indice(_pais, expect, prom):
dicci = {
"Pais": _pais
}
prom, expect = prom[0][2:], expect[0][2:]
indice = [(float(prom[i]) + float(expect[i])) / 2 for i in range(0, len(prom))]
for i in range(1990, 2020):
dicci[str(i)] = indice[i - 1990]
return dicci
for country in promedio["Pais"]:
expect = expectativa.loc[expectativa.Pais == country].to_numpy()
prom = promedio.loc[promedio.Pais == country].to_numpy()
if len(expect) > 0 and len(prom) > 0:
indices = indices.append(calcular_indice(country, expect, prom), ignore_index= True)
```
Resultado del calculo del indice de educción por cada país en cada uno de sus periodos.
```
indices.head()
```
## Almacenamientos de los resultados obtenidos
```
indices.to_csv("Indice_educacion.csv", index=False)
```
|
github_jupyter
|
# Importar modulos necesarios
import pandas as pd
expectativa = pd.read_csv("Expected years of schooling (years).csv", encoding= "latin1")
expectativa
expectativa.shape
expectativa.columns
head = {'HDI Rank':'Ranking',
'Country':'Pais',
'1990':'1990',
'Unnamed: 3':'Indice_Educacion_1990',
'1991':'1991',
'Unnamed: 5':'Indice_Educacion_1991',
'1992':'1992',
'Unnamed: 7':'Indice_Educacion_1992',
'1993':'1993',
'Unnamed: 9':'Indice_Educacion_1993',
'1994':'1994',
'Unnamed: 11':'Indice_Educacion_1994',
'1995':'1995',
'Unnamed: 13':'Indice_Educacion_1995',
'1996':'1996',
'Unnamed: 15':'Indice_Educacion_1996',
'1997':'1997',
'Unnamed: 17':'Indice_Educacion_1997',
'1998':'1998',
'Unnamed: 19':'Indice_Educacion_1998',
'1999':'1999',
'Unnamed: 21':'Indice_Educacion_1999',
'2000':'2000',
'Unnamed: 23':'Indice_Educacion_2000',
'2001':'2001',
'Unnamed: 25':'Indice_Educacion_2001',
'2002':'2002',
'Unnamed: 27':'Indice_Educacion_2002',
'2003':'2003',
'Unnamed: 29':'Indice_Educacion_2003',
'2004':'2004',
'Unnamed: 31':'Indice_Educacion_2004',
'2005':'2005',
'Unnamed: 33':'Indice_Educacion_2005',
'2006':'2006',
'Unnamed: 35':'Indice_Educacion_2006',
'2007':'2007',
'Unnamed: 37':'Indice_Educacion_2007',
'2008':'2008',
'Unnamed: 39':'Indice_Educacion_2008',
'2009':'2009',
'Unnamed: 41':'Indice_Educacion_2009',
'2010':'2010',
'Unnamed: 43':'Indice_Educacion_2010',
'2011':'2011',
'Unnamed: 45':'Indice_Educacion_2011',
'2012':'2012',
'Unnamed: 47':'Indice_Educacion_2012',
'2013':'2013',
'Unnamed: 49':'Indice_Educacion_2013',
'2014':'2014',
'Unnamed: 51':'Indice_Educacion_2014',
'2015':'2015',
'Unnamed: 53':'Indice_Educacion_2015',
'2016':'2016',
'Unnamed: 55':'Indice_Educacion_2016',
'2017':'2017',
'Unnamed: 57':'Indice_Educacion_2017',
'2018':'2018',
'Unnamed: 59':'Indice_Educacion_2018',
'2019':'2019',
'Unnamed: 61':'Indice_Educacion_2019'}
expectativa = expectativa.rename(columns=head)
expectativa.head()
for year in range(1990, 2020):
year = str(year)
indices = []
for cell in expectativa[year]:
try:
indices.append((float(cell) - 0) / (18 - 0))
except:
indices.append(0)
expectativa[f'Indice_Educacion_{year}'] = indices
expectativa = expectativa.drop([str(i) for i in range(1990, 2020)],axis=1)
expectativa.head()
promedio = pd.read_csv("Mean years of schooling (years).csv")
promedio
promedio.shape
promedio.columns
head = {'HDI Rank':'Ranking',
'Country':'Pais',
'1990':'1990',
'Unnamed: 3':'Indice_Promedio_Edu_1990',
'1991':'1991',
'Unnamed: 5':'Indice_Promedio_Edu_1991',
'1992':'1992',
'Unnamed: 7':'Indice_Promedio_Edu_1992',
'1993':'1993',
'Unnamed: 9':'Indice_Promedio_Edu_1993',
'1994':'1994',
'Unnamed: 11':'Indice_Promedio_Edu_1994',
'1995':'1995',
'Unnamed: 13':'Indice_Promedio_Edu_1995',
'1996':'1996',
'Unnamed: 15':'Indice_Promedio_Edu_1996',
'1997':'1997',
'Unnamed: 17':'Indice_Promedio_Edu_1997',
'1998':'1998',
'Unnamed: 19':'Indice_Promedio_Edu_1998',
'1999':'1999',
'Unnamed: 21':'Indice_Promedio_Edu_1999',
'2000':'2000',
'Unnamed: 23':'Indice_Promedio_Edu_2000',
'2001':'2001',
'Unnamed: 25':'Indice_Promedio_Edu_2001',
'2002':'2002',
'Unnamed: 27':'Indice_Promedio_Edu_2002',
'2003':'2003',
'Unnamed: 29':'Indice_Promedio_Edu_2003',
'2004':'2004',
'Unnamed: 31':'Indice_Promedio_Edu_2004',
'2005':'2005',
'Unnamed: 33':'Indice_Promedio_Edu_2005',
'2006':'2006',
'Unnamed: 35':'Indice_Promedio_Edu_2006',
'2007':'2007',
'Unnamed: 37':'Indice_Promedio_Edu_2007',
'2008':'2008',
'Unnamed: 39':'Indice_Promedio_Edu_2008',
'2009':'2009',
'Unnamed: 41':'Indice_Promedio_Edu_2009',
'2010':'2010',
'Unnamed: 43':'Indice_Promedio_Edu_2010',
'2011':'2011',
'Unnamed: 45':'Indice_Promedio_Edu_2011',
'2012':'2012',
'Unnamed: 47':'Indice_Promedio_Edu_2012',
'2013':'2013',
'Unnamed: 49':'Indice_Promedio_Edu_2013',
'2014':'2014',
'Unnamed: 51':'Indice_Promedio_Edu_2014',
'2015':'2015',
'Unnamed: 53':'Indice_Promedio_Edu_2015',
'2016':'2016',
'Unnamed: 55':'Indice_Promedio_Edu_2016',
'2017':'2017',
'Unnamed: 57':'Indice_Promedio_Edu_2017',
'2018':'2018',
'Unnamed: 59':'Indice_Promedio_Edu_2018',
'2019':'2019',
'Unnamed: 61':'Indice_Promedio_Edu_2019'}
promedio = promedio.rename(columns=head)
promedio.head()
import numpy as np
for year in range(1990, 2020):
year = str(year)
indices = []
for cell in promedio[year]:
try:
indices.append((float(cell) - 0) / (15 - 0))
except:
indices.append(0)
promedio[f'Indice_Promedio_Edu_{year}'] = indices
promedio = promedio.drop([str(i) for i in range(1990, 2020)], axis=1)
promedio.head()
header = ['Pais']
indices = pd.DataFrame({}, columns=header)
import numpy as np
def calcular_indice(_pais, expect, prom):
dicci = {
"Pais": _pais
}
prom, expect = prom[0][2:], expect[0][2:]
indice = [(float(prom[i]) + float(expect[i])) / 2 for i in range(0, len(prom))]
for i in range(1990, 2020):
dicci[str(i)] = indice[i - 1990]
return dicci
for country in promedio["Pais"]:
expect = expectativa.loc[expectativa.Pais == country].to_numpy()
prom = promedio.loc[promedio.Pais == country].to_numpy()
if len(expect) > 0 and len(prom) > 0:
indices = indices.append(calcular_indice(country, expect, prom), ignore_index= True)
indices.head()
indices.to_csv("Indice_educacion.csv", index=False)
| 0.358353 | 0.899343 |
```
from lets_plot import *
LetsPlot.setup_html()
```
### Plotting means and error ranges.
There are several ways to show error ranges on a plot. Among them are
- *geom_errorbar*
- *geom_crossbar*
- *geom_linerange*
- *geom_pointrange*
```
# This example was found at: www.cookbook-r.com/Graphs/Plotting_means_and_error_bars_(ggplot2)
data = dict(
supp = ['OJ', 'OJ', 'OJ', 'VC', 'VC', 'VC'],
dose = [0.5, 1.0, 2.0, 0.5, 1.0, 2.0],
length = [13.23, 22.70, 26.06, 7.98, 16.77, 26.14],
len_min = [11.83, 21.2, 24.50, 4.24, 15.26, 23.35],
len_max = [15.63, 24.9, 27.11, 10.72, 19.28, 28.93]
)
p = ggplot(data, aes(x='dose', color='supp'))
```
### Error-bars with lines and points.
```
p + geom_errorbar(aes(ymin='len_min', ymax='len_max'), width=.1) \
+ geom_line(aes(y='length')) \
+ geom_point(aes(y='length'))
# The errorbars overlapped, so use position_dodge to move them horizontally
pd = position_dodge(0.1) # move them .05 to the left and right
p + geom_errorbar(aes(ymin='len_min', ymax='len_max'), width=.1, position=pd) \
+ geom_line(aes(y='length'), position=pd) \
+ geom_point(aes(y='length'), position=pd)
# Black errorbars - notice the mapping of 'group=supp'
# Without it, the errorbars won't be dodged!
p + geom_errorbar(aes(ymin='len_min', ymax='len_max', group='supp'), color='black', width=.1, position=pd) \
+ geom_line(aes(y='length'), position=pd) \
+ geom_point(aes(y='length'), position=pd, size=5)
# The finished graph:
# - fixed size
# - point shape # 21 is filled circle
# - position legend in the bottom right
p1 = p \
+ xlab("Dose (mg)") \
+ ylab("Tooth length (mm)") \
+ scale_color_manual(['orange', 'dark_green'], na_value='gray') \
+ ggsize(700, 400)
p1 + geom_errorbar(aes(ymin='len_min', ymax='len_max', group='supp'), color='black', width=.1, position=pd) \
+ geom_line(aes(y='length'), position=pd) \
+ geom_point(aes(y='length'), position=pd, size=5, shape=21, fill="white") \
+ theme(legend_justification=[1,0], legend_position=[1,0]) \
+ ggtitle("The Effect of Vitamin C on Tooth Growth in Guinea Pigs")
```
### Error-bars on bar plot.
```
# Plot error ranges on Bar plot
p1 \
+ geom_bar(aes(y='length', fill='supp'), stat='identity', position='dodge', color='black') \
+ geom_errorbar(aes(ymin='len_min', ymax='len_max', group='supp'), color='black', width=.1, position=position_dodge(0.9)) \
+ theme(legend_justification=[0,1], legend_position=[0,1])
```
### Crossbars.
```
# Thickness of the horizontal mid-line can be adjusted using `fatten` parameter.
p1 + geom_crossbar(aes(ymin='len_min', ymax='len_max', middle='length', color='supp'), fatten=5)
```
### Line-range.
```
p1 \
+ geom_linerange(aes(ymin='len_min', ymax='len_max', color='supp'), position=pd) \
+ geom_line(aes(y='length'), position=pd)
```
### Point-range
```
# Point-range is the same as line-range but with an added mid-point.
p1 \
+ geom_pointrange(aes(y='length', ymin='len_min', ymax='len_max', color='supp'), position=pd) \
+ geom_line(aes(y='length'), position=pd)
# Size of the mid-point can be adjuasted using `fatten` parameter - multiplication factor relative to the line size.
p1 \
+ geom_line(aes(y='length'), position=pd) \
+ geom_pointrange(aes(y='length', ymin='len_min', ymax='len_max', fill='supp'), position=pd, color='rgb(230, 230, 230)', size=5, shape=23, fatten=1) \
+ scale_fill_manual(['orange', 'dark_green'], na_value='gray')
```
|
github_jupyter
|
from lets_plot import *
LetsPlot.setup_html()
# This example was found at: www.cookbook-r.com/Graphs/Plotting_means_and_error_bars_(ggplot2)
data = dict(
supp = ['OJ', 'OJ', 'OJ', 'VC', 'VC', 'VC'],
dose = [0.5, 1.0, 2.0, 0.5, 1.0, 2.0],
length = [13.23, 22.70, 26.06, 7.98, 16.77, 26.14],
len_min = [11.83, 21.2, 24.50, 4.24, 15.26, 23.35],
len_max = [15.63, 24.9, 27.11, 10.72, 19.28, 28.93]
)
p = ggplot(data, aes(x='dose', color='supp'))
p + geom_errorbar(aes(ymin='len_min', ymax='len_max'), width=.1) \
+ geom_line(aes(y='length')) \
+ geom_point(aes(y='length'))
# The errorbars overlapped, so use position_dodge to move them horizontally
pd = position_dodge(0.1) # move them .05 to the left and right
p + geom_errorbar(aes(ymin='len_min', ymax='len_max'), width=.1, position=pd) \
+ geom_line(aes(y='length'), position=pd) \
+ geom_point(aes(y='length'), position=pd)
# Black errorbars - notice the mapping of 'group=supp'
# Without it, the errorbars won't be dodged!
p + geom_errorbar(aes(ymin='len_min', ymax='len_max', group='supp'), color='black', width=.1, position=pd) \
+ geom_line(aes(y='length'), position=pd) \
+ geom_point(aes(y='length'), position=pd, size=5)
# The finished graph:
# - fixed size
# - point shape # 21 is filled circle
# - position legend in the bottom right
p1 = p \
+ xlab("Dose (mg)") \
+ ylab("Tooth length (mm)") \
+ scale_color_manual(['orange', 'dark_green'], na_value='gray') \
+ ggsize(700, 400)
p1 + geom_errorbar(aes(ymin='len_min', ymax='len_max', group='supp'), color='black', width=.1, position=pd) \
+ geom_line(aes(y='length'), position=pd) \
+ geom_point(aes(y='length'), position=pd, size=5, shape=21, fill="white") \
+ theme(legend_justification=[1,0], legend_position=[1,0]) \
+ ggtitle("The Effect of Vitamin C on Tooth Growth in Guinea Pigs")
# Plot error ranges on Bar plot
p1 \
+ geom_bar(aes(y='length', fill='supp'), stat='identity', position='dodge', color='black') \
+ geom_errorbar(aes(ymin='len_min', ymax='len_max', group='supp'), color='black', width=.1, position=position_dodge(0.9)) \
+ theme(legend_justification=[0,1], legend_position=[0,1])
# Thickness of the horizontal mid-line can be adjusted using `fatten` parameter.
p1 + geom_crossbar(aes(ymin='len_min', ymax='len_max', middle='length', color='supp'), fatten=5)
p1 \
+ geom_linerange(aes(ymin='len_min', ymax='len_max', color='supp'), position=pd) \
+ geom_line(aes(y='length'), position=pd)
# Point-range is the same as line-range but with an added mid-point.
p1 \
+ geom_pointrange(aes(y='length', ymin='len_min', ymax='len_max', color='supp'), position=pd) \
+ geom_line(aes(y='length'), position=pd)
# Size of the mid-point can be adjuasted using `fatten` parameter - multiplication factor relative to the line size.
p1 \
+ geom_line(aes(y='length'), position=pd) \
+ geom_pointrange(aes(y='length', ymin='len_min', ymax='len_max', fill='supp'), position=pd, color='rgb(230, 230, 230)', size=5, shape=23, fatten=1) \
+ scale_fill_manual(['orange', 'dark_green'], na_value='gray')
| 0.881793 | 0.906446 |
```
# A sanity check for the implementation of MADE.
import optax
from jaxrl.networks.policies import NormalTanhMixturePolicy
from jaxrl.networks.autoregressive_policy import MADETanhMixturePolicy
import matplotlib.pyplot as plt
import jax
import numpy as np
import jax.numpy as jnp
import matplotlib
%matplotlib inline
@jax.jit
def sample(rng, inputs, std=0.1):
num_points = len(inputs)
rng, key = jax.random.split(rng)
n = jnp.sqrt(jax.random.uniform(key, shape=(num_points // 2,))
) * 540 * (2 * np.pi) / 360
rng, key = jax.random.split(rng)
d1x = -jnp.cos(n) * n + jax.random.uniform(key,
shape=(num_points // 2,)) * 0.5
rng, key = jax.random.split(rng)
d1y = jnp.sin(n) * n + jax.random.uniform(key,
shape=(num_points // 2,)) * 0.5
x = jnp.concatenate(
[
jnp.stack([d1x, d1y], axis=-1),
jnp.stack([-d1x, -d1y], axis=-1)
]
)
rng, key = jax.random.split(rng)
x = x / 3 + jax.random.normal(key, x.shape) * std
return jnp.clip(x / 5 + inputs, -0.9999, 0.9999)
tmp = sample(jax.random.PRNGKey(1), jnp.zeros((10024, 2)))
x = plt.hist2d(tmp[:, 0], tmp[:, 1], bins=128)
rng = jax.random.PRNGKey(1)
made = NormalTanhMixturePolicy((128, 128), 2) # Fails
made = MADETanhMixturePolicy((128, 128), 2) # Works
rng, key = jax.random.split(rng)
params = made.init(key, jnp.zeros(2))['params']
optim = optax.adamw(3e-4)
optim_state = optim.init(params)
@jax.jit
def train_step(rng, params, optim_state):
rng, key1, key2 = jax.random.split(rng, 3)
xs = jax.random.normal(key1, shape=(1024, 2)) * 0.1
ys = sample(key2, xs)
def loss_fn(params):
dist = made.apply_fn({'params': params}, xs)
log_probs = dist.log_prob(ys)
return -log_probs.mean()
loss_fn(params)
value, grads = jax.value_and_grad(loss_fn)(params)
updates, new_optim_state = optim.update(grads, optim_state, params)
new_params = optax.apply_updates(params, updates)
return value, rng, new_params, new_optim_state
for i in range(100000):
value, rng, params, optim_state = train_step(rng, params, optim_state)
if i % 10000 == 0:
print(value)
@jax.jit
def get_log_probs(xs, ys, params):
dist = made.apply_fn({'params': params}, xs)
return dist.log_prob(ys)
x = jnp.linspace(-0.9, 0.9, 256)
y = jnp.linspace(-0.9, 0.9, 256)
xv, yv = jnp.meshgrid(x, y)
ys = jnp.stack([xv, yv], -1)
xs = jnp.zeros_like(ys) - 0.2
log_probs = get_log_probs(xs, ys, params)
plt.imshow(jnp.exp(log_probs))
```
|
github_jupyter
|
# A sanity check for the implementation of MADE.
import optax
from jaxrl.networks.policies import NormalTanhMixturePolicy
from jaxrl.networks.autoregressive_policy import MADETanhMixturePolicy
import matplotlib.pyplot as plt
import jax
import numpy as np
import jax.numpy as jnp
import matplotlib
%matplotlib inline
@jax.jit
def sample(rng, inputs, std=0.1):
num_points = len(inputs)
rng, key = jax.random.split(rng)
n = jnp.sqrt(jax.random.uniform(key, shape=(num_points // 2,))
) * 540 * (2 * np.pi) / 360
rng, key = jax.random.split(rng)
d1x = -jnp.cos(n) * n + jax.random.uniform(key,
shape=(num_points // 2,)) * 0.5
rng, key = jax.random.split(rng)
d1y = jnp.sin(n) * n + jax.random.uniform(key,
shape=(num_points // 2,)) * 0.5
x = jnp.concatenate(
[
jnp.stack([d1x, d1y], axis=-1),
jnp.stack([-d1x, -d1y], axis=-1)
]
)
rng, key = jax.random.split(rng)
x = x / 3 + jax.random.normal(key, x.shape) * std
return jnp.clip(x / 5 + inputs, -0.9999, 0.9999)
tmp = sample(jax.random.PRNGKey(1), jnp.zeros((10024, 2)))
x = plt.hist2d(tmp[:, 0], tmp[:, 1], bins=128)
rng = jax.random.PRNGKey(1)
made = NormalTanhMixturePolicy((128, 128), 2) # Fails
made = MADETanhMixturePolicy((128, 128), 2) # Works
rng, key = jax.random.split(rng)
params = made.init(key, jnp.zeros(2))['params']
optim = optax.adamw(3e-4)
optim_state = optim.init(params)
@jax.jit
def train_step(rng, params, optim_state):
rng, key1, key2 = jax.random.split(rng, 3)
xs = jax.random.normal(key1, shape=(1024, 2)) * 0.1
ys = sample(key2, xs)
def loss_fn(params):
dist = made.apply_fn({'params': params}, xs)
log_probs = dist.log_prob(ys)
return -log_probs.mean()
loss_fn(params)
value, grads = jax.value_and_grad(loss_fn)(params)
updates, new_optim_state = optim.update(grads, optim_state, params)
new_params = optax.apply_updates(params, updates)
return value, rng, new_params, new_optim_state
for i in range(100000):
value, rng, params, optim_state = train_step(rng, params, optim_state)
if i % 10000 == 0:
print(value)
@jax.jit
def get_log_probs(xs, ys, params):
dist = made.apply_fn({'params': params}, xs)
return dist.log_prob(ys)
x = jnp.linspace(-0.9, 0.9, 256)
y = jnp.linspace(-0.9, 0.9, 256)
xv, yv = jnp.meshgrid(x, y)
ys = jnp.stack([xv, yv], -1)
xs = jnp.zeros_like(ys) - 0.2
log_probs = get_log_probs(xs, ys, params)
plt.imshow(jnp.exp(log_probs))
| 0.492676 | 0.641493 |
A Simple Pytorch Tutorial
======================
PyTorch is a neural network library in Python. Before we dive into PyTorch, let us first think of what functionalities a neural network library should support:
* Implement a neural network $\mathbf{y} = f_\mathbf{\theta}(\mathbf{x})$ parameterized by $\mathbf{\theta}$
* Support Inference: given an input example $\mathbf{x}$, compute the output of the network $\mathbf{y}$
* Support training of the network givan a bunch of training data $\langle \mathbf{x}, \mathbf{y} \rangle$
- The library should support auto-differentiation: computing the gradient $\frac{\partial f_\mathbf{\theta}(\mathbf{x})}{\partial \mathbf{\theta}}$
- The library should also provide implementations of common optimizers (e.g., SGD, Adam) to update the network's parameter $\mathbf{\theta}$ via gradient descent
Pytorch provides receipies to do all of these (and more)!
Let us try to understand some basic concepts of PyTorch using a toy example. Assume the network network we'd like to implement is a simple linear model:
$$ y = \mathbf{w}^\intercal \mathbf{x} + b $$
where our model's parameter $\mathbf{\theta} = \langle \mathbf{w}, b \rangle$.
```
import torch
import torch.nn as nn # where most neural network modules are
```
We could implement our simple neural network by implementing the (abstract) class ``nn.Module``. Basically, what we are going to do is
* Create two model parameters, $\mathbf{w}$ and $b$.
* Define the computation routine of the network: how $y$ is computed given $\mathbf{x}$
```
class MyNeuralNet(nn.Module):
def __init__(self):
super(MyNeuralNet, self).__init__()
# $w$ is 2-dimentional a vector with default value [1.0, 2.0]
self.w = nn.Parameter(
torch.tensor([1.0, 2.0])
)
# b is a scalar with default value 0.5
self.b = nn.Parameter(
torch.tensor(0.5)
)
def forward(self, x):
"""The forward pass"""
y = self.w.dot(x) + self.b
return y
```
Having defined our simple ''neural net'', now let's create one instance and run it!
```
f = MyNeuralNet()
x = torch.tensor([0.5, 0.3])
y = f(x)
print(f'The output `y` is {y}')
```
We then call ``.backward()`` on the output of the nerual network ``y`` to perform back propagation, which compuates the gradients w.r.t model parameters $\mathbf{\theta} = \langle \mathbf{w}, b \rangle$
```
y.backward()
```
To get the gradients of model parameters $\mathbf{\theta} = \langle \mathbf{w}, b \rangle$, simply check the ``.grad`` property of model parameters:
```
print(f'gradient of $w$: {f.w.grad}')
print(f'gradient of $b$: {f.b.grad}')
```
We could do a sanity check to make sure the results are correct:
$$\frac{\partial y}{\partial \mathbf{w}} = \mathbf{x} \quad \frac{\partial y}{\partial b} = 1.0$$
|
github_jupyter
|
import torch
import torch.nn as nn # where most neural network modules are
class MyNeuralNet(nn.Module):
def __init__(self):
super(MyNeuralNet, self).__init__()
# $w$ is 2-dimentional a vector with default value [1.0, 2.0]
self.w = nn.Parameter(
torch.tensor([1.0, 2.0])
)
# b is a scalar with default value 0.5
self.b = nn.Parameter(
torch.tensor(0.5)
)
def forward(self, x):
"""The forward pass"""
y = self.w.dot(x) + self.b
return y
f = MyNeuralNet()
x = torch.tensor([0.5, 0.3])
y = f(x)
print(f'The output `y` is {y}')
y.backward()
print(f'gradient of $w$: {f.w.grad}')
print(f'gradient of $b$: {f.b.grad}')
| 0.922752 | 0.994641 |
```
import sys
sys.path.append("../set-generation")
import csv
import json
import numpy as np
import numba
import color_conversions
ALL_NUM_COLORS = [6, 8, 10]
def to_rgb(color):
"""
Convert hex color code (without `#`) to sRGB255.
"""
return np.array([(int(i[:2], 16), int(i[2:4], 16), int(i[4:], 16)) for i in color])
@numba.njit
def calc_min_dists(rgb):
"""Calculate min delta E for each set."""
nc = rgb.shape[1]
min_dists = []
rgb_linear = color_conversions.sRGB1_to_sRGB1_linear(rgb.flatten()).reshape(
rgb.shape
)
for c in range(rgb_linear.shape[0]):
min_dist = 100
for i in range(1, nc):
for severity in range(1, 101):
jab1 = color_conversions.rgb_linear_to_jab(rgb_linear[c, i])
deut1 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_deuteranomaly(
rgb_linear[c, i], severity
)
)
prot1 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_protanomaly(
rgb_linear[c, i], severity
)
)
trit1 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_tritanomaly(
rgb_linear[c, i], severity
)
)
for j in range(i):
jab2 = color_conversions.rgb_linear_to_jab(rgb_linear[c, j])
deut2 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_deuteranomaly(
rgb_linear[c, j], severity
)
)
prot2 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_protanomaly(
rgb_linear[c, j], severity
)
)
trit2 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_tritanomaly(
rgb_linear[c, j], severity
)
)
min_dist = min(min_dist, color_conversions.cam02de(jab1, jab2))
min_dist = min(min_dist, color_conversions.cam02de(deut1, deut2))
min_dist = min(min_dist, color_conversions.cam02de(prot1, prot2))
min_dist = min(min_dist, color_conversions.cam02de(trit1, trit2))
min_dists.append(min_dist)
return min_dists
```
## With original lightness constraints
```
COLOR_FILE = {
6: "../survey/color-sets/colors_mcd20_mld2_nc6_cvd100_minj40_maxj90_ns10000.txt",
8: "../survey/color-sets/colors_mcd18_mld2_nc8_cvd100_minj40_maxj90_ns10000.txt",
10: "../survey/color-sets/colors_mcd16_mld2_nc10_cvd100_minj40_maxj90_ns10000.txt",
}
# Load color data
colors_rgb = {}
for num_colors in ALL_NUM_COLORS:
with open(COLOR_FILE[num_colors]) as csv_file:
# Skip header rows
csv_file.readline()
csv_file.readline()
csv_file.readline()
csv_reader = csv.reader(csv_file, delimiter=" ")
colors_hex = np.array([[i.strip() for i in row] for row in csv_reader])
colors_rgb[num_colors] = np.array([to_rgb(i) for i in colors_hex]) / 255
min_dists = {nc: np.array(calc_min_dists(colors_rgb[nc])) for nc in ALL_NUM_COLORS}
print("maximum minimum-color distances:")
for nc in ALL_NUM_COLORS:
print(f"{nc:2d}: {np.max(min_dists[nc]):.1f} [{np.argmax(min_dists[nc]):04d}]")
print("mean minimum-color distances:")
for nc in ALL_NUM_COLORS:
print(f"{nc:2d}: {np.mean(min_dists[nc]):.1f}")
```
## With tighter lightness constraints
```
COLOR_FILE = {
6: "../set-generation/colors_mcd20.0_mld5.0_nc6_cvd100_minj40_maxj80_ns10000_f.txt",
8: "../set-generation/colors_mcd18.0_mld4.2_nc8_cvd100_minj40_maxj82_ns10000_f.txt",
10: "../set-generation/colors_mcd16.0_mld3.6_nc10_cvd100_minj40_maxj84_ns10000_f.txt",
}
# Load color data
colors_rgb = {}
colors_hex = {}
for num_colors in ALL_NUM_COLORS:
with open(COLOR_FILE[num_colors]) as csv_file:
# Skip header rows
csv_file.readline()
csv_file.readline()
csv_file.readline()
csv_reader = csv.reader(csv_file, delimiter=" ")
colors_hex[num_colors] = np.array(
[[i.strip() for i in row] for row in csv_reader]
)
colors_rgb[num_colors] = (
np.array([to_rgb(i) for i in colors_hex[num_colors]]) / 255
)
min_dists = {nc: np.array(calc_min_dists(colors_rgb[nc])) for nc in ALL_NUM_COLORS}
print("maximum minimum-color distances:")
max_min_dist_sets = {}
for nc in ALL_NUM_COLORS:
max_min_dist_sets[nc] = list(colors_hex[nc][np.argmax(min_dists[nc])])
print(f"{nc:2d}: {np.max(min_dists[nc]):.1f} [{np.argmax(min_dists[nc]):04d}]")
print("mean minimum-color distances:")
for nc in ALL_NUM_COLORS:
print(f"{nc:2d}: {np.mean(min_dists[nc]):.1f}")
with open("max-min-dist-sets.json", "w") as outfile:
json.dump(max_min_dist_sets, outfile)
```
|
github_jupyter
|
import sys
sys.path.append("../set-generation")
import csv
import json
import numpy as np
import numba
import color_conversions
ALL_NUM_COLORS = [6, 8, 10]
def to_rgb(color):
"""
Convert hex color code (without `#`) to sRGB255.
"""
return np.array([(int(i[:2], 16), int(i[2:4], 16), int(i[4:], 16)) for i in color])
@numba.njit
def calc_min_dists(rgb):
"""Calculate min delta E for each set."""
nc = rgb.shape[1]
min_dists = []
rgb_linear = color_conversions.sRGB1_to_sRGB1_linear(rgb.flatten()).reshape(
rgb.shape
)
for c in range(rgb_linear.shape[0]):
min_dist = 100
for i in range(1, nc):
for severity in range(1, 101):
jab1 = color_conversions.rgb_linear_to_jab(rgb_linear[c, i])
deut1 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_deuteranomaly(
rgb_linear[c, i], severity
)
)
prot1 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_protanomaly(
rgb_linear[c, i], severity
)
)
trit1 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_tritanomaly(
rgb_linear[c, i], severity
)
)
for j in range(i):
jab2 = color_conversions.rgb_linear_to_jab(rgb_linear[c, j])
deut2 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_deuteranomaly(
rgb_linear[c, j], severity
)
)
prot2 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_protanomaly(
rgb_linear[c, j], severity
)
)
trit2 = color_conversions.rgb_linear_to_jab(
color_conversions.CVD_forward_tritanomaly(
rgb_linear[c, j], severity
)
)
min_dist = min(min_dist, color_conversions.cam02de(jab1, jab2))
min_dist = min(min_dist, color_conversions.cam02de(deut1, deut2))
min_dist = min(min_dist, color_conversions.cam02de(prot1, prot2))
min_dist = min(min_dist, color_conversions.cam02de(trit1, trit2))
min_dists.append(min_dist)
return min_dists
COLOR_FILE = {
6: "../survey/color-sets/colors_mcd20_mld2_nc6_cvd100_minj40_maxj90_ns10000.txt",
8: "../survey/color-sets/colors_mcd18_mld2_nc8_cvd100_minj40_maxj90_ns10000.txt",
10: "../survey/color-sets/colors_mcd16_mld2_nc10_cvd100_minj40_maxj90_ns10000.txt",
}
# Load color data
colors_rgb = {}
for num_colors in ALL_NUM_COLORS:
with open(COLOR_FILE[num_colors]) as csv_file:
# Skip header rows
csv_file.readline()
csv_file.readline()
csv_file.readline()
csv_reader = csv.reader(csv_file, delimiter=" ")
colors_hex = np.array([[i.strip() for i in row] for row in csv_reader])
colors_rgb[num_colors] = np.array([to_rgb(i) for i in colors_hex]) / 255
min_dists = {nc: np.array(calc_min_dists(colors_rgb[nc])) for nc in ALL_NUM_COLORS}
print("maximum minimum-color distances:")
for nc in ALL_NUM_COLORS:
print(f"{nc:2d}: {np.max(min_dists[nc]):.1f} [{np.argmax(min_dists[nc]):04d}]")
print("mean minimum-color distances:")
for nc in ALL_NUM_COLORS:
print(f"{nc:2d}: {np.mean(min_dists[nc]):.1f}")
COLOR_FILE = {
6: "../set-generation/colors_mcd20.0_mld5.0_nc6_cvd100_minj40_maxj80_ns10000_f.txt",
8: "../set-generation/colors_mcd18.0_mld4.2_nc8_cvd100_minj40_maxj82_ns10000_f.txt",
10: "../set-generation/colors_mcd16.0_mld3.6_nc10_cvd100_minj40_maxj84_ns10000_f.txt",
}
# Load color data
colors_rgb = {}
colors_hex = {}
for num_colors in ALL_NUM_COLORS:
with open(COLOR_FILE[num_colors]) as csv_file:
# Skip header rows
csv_file.readline()
csv_file.readline()
csv_file.readline()
csv_reader = csv.reader(csv_file, delimiter=" ")
colors_hex[num_colors] = np.array(
[[i.strip() for i in row] for row in csv_reader]
)
colors_rgb[num_colors] = (
np.array([to_rgb(i) for i in colors_hex[num_colors]]) / 255
)
min_dists = {nc: np.array(calc_min_dists(colors_rgb[nc])) for nc in ALL_NUM_COLORS}
print("maximum minimum-color distances:")
max_min_dist_sets = {}
for nc in ALL_NUM_COLORS:
max_min_dist_sets[nc] = list(colors_hex[nc][np.argmax(min_dists[nc])])
print(f"{nc:2d}: {np.max(min_dists[nc]):.1f} [{np.argmax(min_dists[nc]):04d}]")
print("mean minimum-color distances:")
for nc in ALL_NUM_COLORS:
print(f"{nc:2d}: {np.mean(min_dists[nc]):.1f}")
with open("max-min-dist-sets.json", "w") as outfile:
json.dump(max_min_dist_sets, outfile)
| 0.572245 | 0.565119 |
# Homework 6 - Liberatori Benedetta
The following is the implementation of a U-Net-style CNN s.t. :
1. All convolutions must use a $3\times 3$ kernel and leave the spatial dimensions of the input untouched.
2. Downsampling in the contracting part is performed via maxpooling with a $2\times 2$ kernel and stride of 2.
3. Upsampling is operated by a deconvolution with a $2\times 2$ kernel and stride of 2.
4. The final layer of the expanding part has only 1 channel
Following the one proposed in [1](https://arxiv.org/abs/1505.04597).

```
import torch
from torch import nn
from torch.utils.data import DataLoader
import torchvision.transforms as T
from torchvision.datasets import CIFAR10
from torchvision.models import vgg11_bn
from torchsummary import summary
from scripts import mnistm
from scripts import mnist
from scripts import train
from matplotlib import pyplot as plt
import numpy as np
```
Both the contracting path (left side) and expansive path (right side) consists of repeated applications of two $3\times 3$ convolutions, each followed by a ReLU activation function.
In the original paper those are unpadded convolutions, this implementation instead is meant to leave the spatial dimensions of the input untouched.
Let us first define a class for this basic unit:
```
class DoubleConv(nn.Module):
def __init__(self, in_channels, out_channels):
super().__init__()
self.features =nn.Sequential(
nn.Conv2d(in_channels, out_channels, kernel_size=3, padding=1),
nn.ReLU(),
nn.Conv2d(out_channels, out_channels, kernel_size=3, padding=1),
nn.ReLU()
)
def forward(self, x):
return self.features(x)
unit = DoubleConv(1, 64)
x = torch.randn(1, 1, 572, 572)
unit(x).shape
```
As expected, the output is a tensor with 64 channels and unchanged spatial dimentions.
Now these briks will be used inside a class for the first half of the network. Between those there will be a $2\times 2$ max pooling operation with stride $2$ for downsampling, doubling the number of feature channels at each step.
torch.nn.ModuleList() holds submodules in a list and can be indexed like a rebular Python list.
The output from the last convolutional layer will be concatenated with the output of upsapling of the corresponding block in the expansive path. Thus these outputs are stored in a list.
```
class Down(nn.Module):
def __init__(self, channels=(1,64,128,256,512,1024)):
super().__init__()
self.conv = nn.ModuleList([DoubleConv(channels[i], channels[i+1]) for i in range(len(channels)-1)])
self.pool = nn.MaxPool2d(2)
def forward(self, x):
out = []
for conv in self.conv:
x = conv(x)
out.append(x)
x = self.pool(x)
return out
```
Now let us check the correctness of dimenstions on random data. The number of feature channels should be doubled at each step and the spatial dimensions should decrease accordingly with the pooling arithmetic rule :
$$ o = \Bigl\lfloor\frac{i-k}{s}\Bigr\rfloor+1$$
where $i$ is the input dimention, $k$ is the kernel size, $s$ the stride and $o$ is the output dimention after pooling. Being our input a square image we just need to check one dimention.
```
def out_dim_pooling(i, k, s, t):
for _ in range(t):
print(i)
i=np.floor((i-k)/s) +1
out_dim_pooling(572, 2, 2, 5)
downpath = Down()
y = downpath(x)
for _ in y: print(_.shape)
```
So the dimentions are as expected.
The following class implements the second half of the architecture, the expansive path.
Each step is:
- a $2\times2$ deconvolution that halves the number of feature channels,
- a concatenation with the correspondingly cropped feature map from the contracting path,
- and two 3x3 convolutions, each followed by a ReLU (basic unit).
For this purpose, the forward method takes in input the features to be concatenated.
torchvision.transforms.CenterCrop([h,w]) is used to crops the given image at the center, suing the dims passed in input. This is needed to ensure that the two parts to be concatenated have the same spatial dimentions.
N.B: In principle, if the input image dimensions are divisible by $2^4$ then this step is not needed since we are using padding in the $3\times 3 $ convolutions.
```
class Up(nn.Module):
def __init__(self, channels=(1024, 512, 256, 128, 64)):
super().__init__()
self.channels=channels
self.upconv= nn.ModuleList([nn.ConvTranspose2d(channels[i], channels[i+1], 2, 2) for i in range(len(channels)-1)])
self.conv= nn.ModuleList([DoubleConv(channels[i], channels[i+1]) for i in range(len(channels)-1)])
def forward(self, x, features_to_concatenate):
for i in range(len(self.channels)-1):
x = self.upconv[i](x)
_,_, h, w = x.shape
cropped = T.CenterCrop([h,w])(features_to_concatenate[i])
x = torch.cat([x, cropped], dim=1)
x = self.conv[i](x)
return x
```
We expect the number of features channels to be $1024/2^4=64$ and the spacial dimentions to be $35\times 2^4=560$.
The tensor y = downpath(x) from before stores the outputs of the last convolutional layer at each block, now it is time to use those. Actually, the last output of size $[1, 1024, 35, 35]$ does not need to be concatenated, and thus to be passed to the Up block.
```
uppath = Up()
x = torch.randn(1, 1024, 35, 35)
uppath(x, y[::-1][1:]).shape
class UNET(nn.Module):
def __init__(self, in_channels, num_classes, d_channels=(1,64,128,256,512,1024), u_channels=(1024, 512, 256, 128, 64)):
super().__init__()
self.contract = Down(d_channels)
self.expand = Up(u_channels)
self.last = nn.Conv2d(64, num_classes, 1)
def forward(self,x):
contr = self.contract(x)
out = self.expand(contr[::-1][0], contr[::-1][1:])
out = self.last(out)
return out
unet = UNET(in_channels=1, num_classes=1)
x = torch.randn(1, 1, 572, 572)
unet(x).shape
```
At the final layer a $1\times1$ convolution is used to map each 64-component feature vector to the desired number of classes. With 1 channel output this is a binary classification at pixel level.
[1](https://arxiv.org/abs/1505.04597) Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention.
|
github_jupyter
|
import torch
from torch import nn
from torch.utils.data import DataLoader
import torchvision.transforms as T
from torchvision.datasets import CIFAR10
from torchvision.models import vgg11_bn
from torchsummary import summary
from scripts import mnistm
from scripts import mnist
from scripts import train
from matplotlib import pyplot as plt
import numpy as np
class DoubleConv(nn.Module):
def __init__(self, in_channels, out_channels):
super().__init__()
self.features =nn.Sequential(
nn.Conv2d(in_channels, out_channels, kernel_size=3, padding=1),
nn.ReLU(),
nn.Conv2d(out_channels, out_channels, kernel_size=3, padding=1),
nn.ReLU()
)
def forward(self, x):
return self.features(x)
unit = DoubleConv(1, 64)
x = torch.randn(1, 1, 572, 572)
unit(x).shape
class Down(nn.Module):
def __init__(self, channels=(1,64,128,256,512,1024)):
super().__init__()
self.conv = nn.ModuleList([DoubleConv(channels[i], channels[i+1]) for i in range(len(channels)-1)])
self.pool = nn.MaxPool2d(2)
def forward(self, x):
out = []
for conv in self.conv:
x = conv(x)
out.append(x)
x = self.pool(x)
return out
def out_dim_pooling(i, k, s, t):
for _ in range(t):
print(i)
i=np.floor((i-k)/s) +1
out_dim_pooling(572, 2, 2, 5)
downpath = Down()
y = downpath(x)
for _ in y: print(_.shape)
class Up(nn.Module):
def __init__(self, channels=(1024, 512, 256, 128, 64)):
super().__init__()
self.channels=channels
self.upconv= nn.ModuleList([nn.ConvTranspose2d(channels[i], channels[i+1], 2, 2) for i in range(len(channels)-1)])
self.conv= nn.ModuleList([DoubleConv(channels[i], channels[i+1]) for i in range(len(channels)-1)])
def forward(self, x, features_to_concatenate):
for i in range(len(self.channels)-1):
x = self.upconv[i](x)
_,_, h, w = x.shape
cropped = T.CenterCrop([h,w])(features_to_concatenate[i])
x = torch.cat([x, cropped], dim=1)
x = self.conv[i](x)
return x
uppath = Up()
x = torch.randn(1, 1024, 35, 35)
uppath(x, y[::-1][1:]).shape
class UNET(nn.Module):
def __init__(self, in_channels, num_classes, d_channels=(1,64,128,256,512,1024), u_channels=(1024, 512, 256, 128, 64)):
super().__init__()
self.contract = Down(d_channels)
self.expand = Up(u_channels)
self.last = nn.Conv2d(64, num_classes, 1)
def forward(self,x):
contr = self.contract(x)
out = self.expand(contr[::-1][0], contr[::-1][1:])
out = self.last(out)
return out
unet = UNET(in_channels=1, num_classes=1)
x = torch.randn(1, 1, 572, 572)
unet(x).shape
| 0.928741 | 0.979861 |
# TensorFlow2.0教程-注意力神经机器翻译
训练序列到序列(seq2seq)模型西班牙语到英语翻译。这是一个高级示例,假定序列模型的序列知识。
在本教程中能够输入西班牙语句子,例如“¿todavia estan en casa?” ,并返回英文翻译:“你还在家吗?”
对于玩具示例而言,翻译质量是合理的,但生成的关注图可能更有趣。这表明输入句子的哪些部分在翻译时具有模型的注意力:
<img src="https://tensorflow.org/images/spanish-english.png" alt="spanish-english attention plot">
```
from __future__ import absolute_import, division, print_function, unicode_literals
!pip install tensorflow-gpu==2.0.0-beta1
import tensorflow as tf
print(tf.__version__)
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
from sklearn.model_selection import train_test_split
import unicodedata
import re
import numpy as np
import os
import io
import time
```
## 下载并准备数据集
我们将使用http://www.manythings.org/anki/提供的语言数据集。此数据集包含以下格式的语言翻译对:
May I borrow this book? ¿Puedo tomar prestado este libro?
有多种语言可供选择,但我们将使用英语 - 西班牙语数据集。为方便起见,我们在Google Cloud上托管了此数据集的副本,但您也可以下载自己的副本。下载数据集后,以下是我们准备数据的步骤:
- 为每个句子添加开始和结束标记。
- 删除特殊字符来清除句子。
- 创建一个单词索引和反向单词索引(从单词→id和id→单词映射的字典)。
- 将每个句子填充到最大长度。
```
# Download the file
path_to_zip = tf.keras.utils.get_file(
'spa-eng.zip', origin='http://storage.googleapis.com/download.tensorflow.org/data/spa-eng.zip',
extract=True)
path_to_file = os.path.dirname(path_to_zip)+"/spa-eng/spa.txt"
# Converts the unicode file to ascii
def unicode_to_ascii(s):
return ''.join(c for c in unicodedata.normalize('NFD', s)
if unicodedata.category(c) != 'Mn')
def preprocess_sentence(w):
w = unicode_to_ascii(w.lower().strip())
# creating a space between a word and the punctuation following it
# eg: "he is a boy." => "he is a boy ."
# Reference:- https://stackoverflow.com/questions/3645931/python-padding-punctuation-with-white-spaces-keeping-punctuation
w = re.sub(r"([?.!,¿])", r" \1 ", w)
w = re.sub(r'[" "]+', " ", w)
# replacing everything with space except (a-z, A-Z, ".", "?", "!", ",")
w = re.sub(r"[^a-zA-Z?.!,¿]+", " ", w)
w = w.rstrip().strip()
# adding a start and an end token to the sentence
# so that the model know when to start and stop predicting.
w = '<start> ' + w + ' <end>'
return w
en_sentence = u"May I borrow this book?"
sp_sentence = u"¿Puedo tomar prestado este libro?"
print(preprocess_sentence(en_sentence))
print(preprocess_sentence(sp_sentence).encode('utf-8'))
# 1. Remove the accents
# 2. Clean the sentences
# 3. Return word pairs in the format: [ENGLISH, SPANISH]
def create_dataset(path, num_examples):
lines = io.open(path, encoding='UTF-8').read().strip().split('\n')
word_pairs = [[preprocess_sentence(w) for w in l.split('\t')] for l in lines[:num_examples]]
return zip(*word_pairs)
en, sp = create_dataset(path_to_file, None)
print(en[-1])
print(sp[-1])
def max_length(tensor):
return max(len(t) for t in tensor)
def tokenize(lang):
lang_tokenizer = tf.keras.preprocessing.text.Tokenizer(
filters='')
lang_tokenizer.fit_on_texts(lang)
tensor = lang_tokenizer.texts_to_sequences(lang)
tensor = tf.keras.preprocessing.sequence.pad_sequences(tensor,
padding='post')
return tensor, lang_tokenizer
def load_dataset(path, num_examples=None):
# creating cleaned input, output pairs
targ_lang, inp_lang = create_dataset(path, num_examples)
input_tensor, inp_lang_tokenizer = tokenize(inp_lang)
target_tensor, targ_lang_tokenizer = tokenize(targ_lang)
return input_tensor, target_tensor, inp_lang_tokenizer, targ_lang_tokenizer
```
### 限制数据集的大小以更快地进行实验(可选)
对> 100,000个句子的完整数据集进行培训需要很长时间。为了更快地训练,我们可以将数据集的大小限制为30,000个句子(当然,翻译质量会随着数据的减少而降低):
```
# Try experimenting with the size of that dataset
num_examples = 30000
input_tensor, target_tensor, inp_lang, targ_lang = load_dataset(path_to_file, num_examples)
# Calculate max_length of the target tensors
max_length_targ, max_length_inp = max_length(target_tensor), max_length(input_tensor)
# Creating training and validation sets using an 80-20 split
input_tensor_train, input_tensor_val, target_tensor_train, target_tensor_val = train_test_split(input_tensor, target_tensor, test_size=0.2)
# Show length
print(len(input_tensor_train), len(target_tensor_train), len(input_tensor_val), len(target_tensor_val))
def convert(lang, tensor):
for t in tensor:
if t!=0:
print ("%d ----> %s" % (t, lang.index_word[t]))
print ("Input Language; index to word mapping")
convert(inp_lang, input_tensor_train[0])
print ()
print ("Target Language; index to word mapping")
convert(targ_lang, target_tensor_train[0])
```
### 创建tf.data数据集
```
BUFFER_SIZE = len(input_tensor_train)
BATCH_SIZE = 64
steps_per_epoch = len(input_tensor_train)//BATCH_SIZE
embedding_dim = 256
units = 1024
vocab_inp_size = len(inp_lang.word_index)+1
vocab_tar_size = len(targ_lang.word_index)+1
dataset = tf.data.Dataset.from_tensor_slices((input_tensor_train, target_tensor_train)).shuffle(BUFFER_SIZE)
dataset = dataset.batch(BATCH_SIZE, drop_remainder=True)
example_input_batch, example_target_batch = next(iter(dataset))
example_input_batch.shape, example_target_batch.shape
```
编写编码器和解码器模型
实现编码器 - 解码器模型,您可以在TensorFlow 神经机器翻译(seq2seq)教程中阅读。此示例使用更新的API集。这个笔记本实现了seq2seq教程中的注意方程式。下图显示了每个输入单词由注意机制分配权重,然后解码器使用该权重来预测句子中的下一个单词。下面的图片和公式是Luong的论文中关注机制的一个例子。
<img src="https://www.tensorflow.org/images/seq2seq/attention_mechanism.jpg" width="500" alt="attention mechanism">
The input is put through an encoder model which gives us the encoder output of shape *(batch_size, max_length, hidden_size)* and the encoder hidden state of shape *(batch_size, hidden_size)*.
Here are the equations that are implemented:
<img src="https://www.tensorflow.org/images/seq2seq/attention_equation_0.jpg" alt="attention equation 0" width="800">
<img src="https://www.tensorflow.org/images/seq2seq/attention_equation_1.jpg" alt="attention equation 1" width="800">
本教程使用Bahdanau注意编码器。在编写简化表单之前,让我们决定表示法:
- FC =完全连接(密集)层
- EO =编码器输出
- H =隐藏状态
- X =解码器的输入
和伪代码:
- score = FC(tanh(FC(EO) + FC(H)))
- attention weights = softmax(score, axis = 1)。默认情况下,Softmax应用于最后一个轴,但是我们要在第一个轴上应用它,因为得分的形状是(batch_size,max_length,hidden_size)。Max_length是我们输入的长度。由于我们尝试为每个输入分配权重,因此应在该轴上应用softmax。
- context vector = sum(attention weights * EO, axis = 1)。选择轴为1的原因与上述相同。
- embedding output =解码器X的输入通过嵌入层。
- merged vector = concat(embedding output, context vector)
- 然后将该合并的矢量提供给GRU
每个步骤中所有向量的形状都已在代码中的注释中指定:
```
class Encoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, enc_units, batch_sz):
super(Encoder, self).__init__()
self.batch_sz = batch_sz
self.enc_units = enc_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = tf.keras.layers.GRU(self.enc_units,
return_sequences=True,
return_state=True,
recurrent_initializer='glorot_uniform')
def call(self, x, hidden):
x = self.embedding(x)
output, state = self.gru(x, initial_state = hidden)
return output, state
def initialize_hidden_state(self):
return tf.zeros((self.batch_sz, self.enc_units))
encoder = Encoder(vocab_inp_size, embedding_dim, units, BATCH_SIZE)
# sample input
sample_hidden = encoder.initialize_hidden_state()
sample_output, sample_hidden = encoder(example_input_batch, sample_hidden)
print ('Encoder output shape: (batch size, sequence length, units) {}'.format(sample_output.shape))
print ('Encoder Hidden state shape: (batch size, units) {}'.format(sample_hidden.shape))
class BahdanauAttention(tf.keras.Model):
def __init__(self, units):
super(BahdanauAttention, self).__init__()
self.W1 = tf.keras.layers.Dense(units)
self.W2 = tf.keras.layers.Dense(units)
self.V = tf.keras.layers.Dense(1)
def call(self, query, values):
# hidden shape == (batch_size, hidden size)
# hidden_with_time_axis shape == (batch_size, 1, hidden size)
# we are doing this to perform addition to calculate the score
hidden_with_time_axis = tf.expand_dims(query, 1)
# score shape == (batch_size, max_length, 1)
# we get 1 at the last axis because we are applying score to self.V
# the shape of the tensor before applying self.V is (batch_size, max_length, units)
score = self.V(tf.nn.tanh(
self.W1(values) + self.W2(hidden_with_time_axis)))
# attention_weights shape == (batch_size, max_length, 1)
attention_weights = tf.nn.softmax(score, axis=1)
# context_vector shape after sum == (batch_size, hidden_size)
context_vector = attention_weights * values
context_vector = tf.reduce_sum(context_vector, axis=1)
return context_vector, attention_weights
attention_layer = BahdanauAttention(10)
attention_result, attention_weights = attention_layer(sample_hidden, sample_output)
print("Attention result shape: (batch size, units) {}".format(attention_result.shape))
print("Attention weights shape: (batch_size, sequence_length, 1) {}".format(attention_weights.shape))
class Decoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, dec_units, batch_sz):
super(Decoder, self).__init__()
self.batch_sz = batch_sz
self.dec_units = dec_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = tf.keras.layers.GRU(self.dec_units,
return_sequences=True,
return_state=True,
recurrent_initializer='glorot_uniform')
self.fc = tf.keras.layers.Dense(vocab_size)
# used for attention
self.attention = BahdanauAttention(self.dec_units)
def call(self, x, hidden, enc_output):
# enc_output shape == (batch_size, max_length, hidden_size)
context_vector, attention_weights = self.attention(hidden, enc_output)
# x shape after passing through embedding == (batch_size, 1, embedding_dim)
x = self.embedding(x)
# x shape after concatenation == (batch_size, 1, embedding_dim + hidden_size)
x = tf.concat([tf.expand_dims(context_vector, 1), x], axis=-1)
# passing the concatenated vector to the GRU
output, state = self.gru(x)
# output shape == (batch_size * 1, hidden_size)
output = tf.reshape(output, (-1, output.shape[2]))
# output shape == (batch_size, vocab)
x = self.fc(output)
return x, state, attention_weights
decoder = Decoder(vocab_tar_size, embedding_dim, units, BATCH_SIZE)
sample_decoder_output, _, _ = decoder(tf.random.uniform((64, 1)),
sample_hidden, sample_output)
print ('Decoder output shape: (batch_size, vocab size) {}'.format(sample_decoder_output.shape))
```
## 定义优化器和损失函数
```
optimizer = tf.keras.optimizers.Adam()
loss_object = tf.keras.losses.SparseCategoricalCrossentropy(
from_logits=True, reduction='none')
def loss_function(real, pred):
mask = tf.math.logical_not(tf.math.equal(real, 0))
loss_ = loss_object(real, pred)
mask = tf.cast(mask, dtype=loss_.dtype)
loss_ *= mask
return tf.reduce_mean(loss_)
```
## 检查点(基于对象的保存)
```
checkpoint_dir = './training_checkpoints'
checkpoint_prefix = os.path.join(checkpoint_dir, "ckpt")
checkpoint = tf.train.Checkpoint(optimizer=optimizer,
encoder=encoder,
decoder=decoder)
```
## 训练
- 通过编码器传递输入,编码器返回编码器输出和编码器隐藏状态。
- 编码器输出,编码器隐藏状态和解码器输入(它是开始标记)被传递给解码器。
- 解码器返回预测和解码器隐藏状态。
- 然后将解码器隐藏状态传递回模型,并使用预测来计算损失。
- 使用教师强制决定解码器的下一个输入。
- 教师强制是将目标字作为下一个输入传递给解码器的技术。
- 最后一步是计算渐变并将其应用于优化器并反向传播。
```
@tf.function
def train_step(inp, targ, enc_hidden):
loss = 0
with tf.GradientTape() as tape:
enc_output, enc_hidden = encoder(inp, enc_hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word_index['<start>']] * BATCH_SIZE, 1)
# Teacher forcing - feeding the target as the next input
for t in range(1, targ.shape[1]):
# passing enc_output to the decoder
predictions, dec_hidden, _ = decoder(dec_input, dec_hidden, enc_output)
loss += loss_function(targ[:, t], predictions)
# using teacher forcing
dec_input = tf.expand_dims(targ[:, t], 1)
batch_loss = (loss / int(targ.shape[1]))
variables = encoder.trainable_variables + decoder.trainable_variables
gradients = tape.gradient(loss, variables)
optimizer.apply_gradients(zip(gradients, variables))
return batch_loss
EPOCHS = 10
for epoch in range(EPOCHS):
start = time.time()
enc_hidden = encoder.initialize_hidden_state()
total_loss = 0
for (batch, (inp, targ)) in enumerate(dataset.take(steps_per_epoch)):
batch_loss = train_step(inp, targ, enc_hidden)
total_loss += batch_loss
if batch % 100 == 0:
print('Epoch {} Batch {} Loss {:.4f}'.format(epoch + 1,
batch,
batch_loss.numpy()))
# saving (checkpoint) the model every 2 epochs
if (epoch + 1) % 2 == 0:
checkpoint.save(file_prefix = checkpoint_prefix)
print('Epoch {} Loss {:.4f}'.format(epoch + 1,
total_loss / steps_per_epoch))
print('Time taken for 1 epoch {} sec\n'.format(time.time() - start))
```
## 翻译
- evaluate函数类似于训练循环,除了我们在这里不使用教师强制。在每个时间步骤对解码器的输入是其先前的预测以及隐藏状态和编码器输出。
- 停止预测模型何时预测结束标记。
- 并存储每个时间步的注意力。
注意:编码器输出仅针对一个输入计算一次。
```
def evaluate(sentence):
attention_plot = np.zeros((max_length_targ, max_length_inp))
sentence = preprocess_sentence(sentence)
inputs = [inp_lang.word_index[i] for i in sentence.split(' ')]
inputs = tf.keras.preprocessing.sequence.pad_sequences([inputs],
maxlen=max_length_inp,
padding='post')
inputs = tf.convert_to_tensor(inputs)
result = ''
hidden = [tf.zeros((1, units))]
enc_out, enc_hidden = encoder(inputs, hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word_index['<start>']], 0)
for t in range(max_length_targ):
predictions, dec_hidden, attention_weights = decoder(dec_input,
dec_hidden,
enc_out)
# storing the attention weights to plot later on
attention_weights = tf.reshape(attention_weights, (-1, ))
attention_plot[t] = attention_weights.numpy()
predicted_id = tf.argmax(predictions[0]).numpy()
result += targ_lang.index_word[predicted_id] + ' '
if targ_lang.index_word[predicted_id] == '<end>':
return result, sentence, attention_plot
# the predicted ID is fed back into the model
dec_input = tf.expand_dims([predicted_id], 0)
return result, sentence, attention_plot
# function for plotting the attention weights
def plot_attention(attention, sentence, predicted_sentence):
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot(1, 1, 1)
ax.matshow(attention, cmap='viridis')
fontdict = {'fontsize': 14}
ax.set_xticklabels([''] + sentence, fontdict=fontdict, rotation=90)
ax.set_yticklabels([''] + predicted_sentence, fontdict=fontdict)
ax.xaxis.set_major_locator(ticker.MultipleLocator(1))
ax.yaxis.set_major_locator(ticker.MultipleLocator(1))
plt.show()
def translate(sentence):
result, sentence, attention_plot = evaluate(sentence)
print('Input: %s' % (sentence))
print('Predicted translation: {}'.format(result))
attention_plot = attention_plot[:len(result.split(' ')), :len(sentence.split(' '))]
plot_attention(attention_plot, sentence.split(' '), result.split(' '))
```
## 恢复最新的检查点并进行测试
```
# restoring the latest checkpoint in checkpoint_dir
checkpoint.restore(tf.train.latest_checkpoint(checkpoint_dir))
translate(u'hace mucho frio aqui.')
translate(u'esta es mi vida.')
translate(u'¿todavia estan en casa?')
# wrong translation
translate(u'trata de averiguarlo.')
```
## 下一步
- 下载不同的数据集以试验翻译,例如,英语到德语,或英语到法语。
- 尝试对更大的数据集进行训练,或使用更多的时期
|
github_jupyter
|
from __future__ import absolute_import, division, print_function, unicode_literals
!pip install tensorflow-gpu==2.0.0-beta1
import tensorflow as tf
print(tf.__version__)
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
from sklearn.model_selection import train_test_split
import unicodedata
import re
import numpy as np
import os
import io
import time
# Download the file
path_to_zip = tf.keras.utils.get_file(
'spa-eng.zip', origin='http://storage.googleapis.com/download.tensorflow.org/data/spa-eng.zip',
extract=True)
path_to_file = os.path.dirname(path_to_zip)+"/spa-eng/spa.txt"
# Converts the unicode file to ascii
def unicode_to_ascii(s):
return ''.join(c for c in unicodedata.normalize('NFD', s)
if unicodedata.category(c) != 'Mn')
def preprocess_sentence(w):
w = unicode_to_ascii(w.lower().strip())
# creating a space between a word and the punctuation following it
# eg: "he is a boy." => "he is a boy ."
# Reference:- https://stackoverflow.com/questions/3645931/python-padding-punctuation-with-white-spaces-keeping-punctuation
w = re.sub(r"([?.!,¿])", r" \1 ", w)
w = re.sub(r'[" "]+', " ", w)
# replacing everything with space except (a-z, A-Z, ".", "?", "!", ",")
w = re.sub(r"[^a-zA-Z?.!,¿]+", " ", w)
w = w.rstrip().strip()
# adding a start and an end token to the sentence
# so that the model know when to start and stop predicting.
w = '<start> ' + w + ' <end>'
return w
en_sentence = u"May I borrow this book?"
sp_sentence = u"¿Puedo tomar prestado este libro?"
print(preprocess_sentence(en_sentence))
print(preprocess_sentence(sp_sentence).encode('utf-8'))
# 1. Remove the accents
# 2. Clean the sentences
# 3. Return word pairs in the format: [ENGLISH, SPANISH]
def create_dataset(path, num_examples):
lines = io.open(path, encoding='UTF-8').read().strip().split('\n')
word_pairs = [[preprocess_sentence(w) for w in l.split('\t')] for l in lines[:num_examples]]
return zip(*word_pairs)
en, sp = create_dataset(path_to_file, None)
print(en[-1])
print(sp[-1])
def max_length(tensor):
return max(len(t) for t in tensor)
def tokenize(lang):
lang_tokenizer = tf.keras.preprocessing.text.Tokenizer(
filters='')
lang_tokenizer.fit_on_texts(lang)
tensor = lang_tokenizer.texts_to_sequences(lang)
tensor = tf.keras.preprocessing.sequence.pad_sequences(tensor,
padding='post')
return tensor, lang_tokenizer
def load_dataset(path, num_examples=None):
# creating cleaned input, output pairs
targ_lang, inp_lang = create_dataset(path, num_examples)
input_tensor, inp_lang_tokenizer = tokenize(inp_lang)
target_tensor, targ_lang_tokenizer = tokenize(targ_lang)
return input_tensor, target_tensor, inp_lang_tokenizer, targ_lang_tokenizer
# Try experimenting with the size of that dataset
num_examples = 30000
input_tensor, target_tensor, inp_lang, targ_lang = load_dataset(path_to_file, num_examples)
# Calculate max_length of the target tensors
max_length_targ, max_length_inp = max_length(target_tensor), max_length(input_tensor)
# Creating training and validation sets using an 80-20 split
input_tensor_train, input_tensor_val, target_tensor_train, target_tensor_val = train_test_split(input_tensor, target_tensor, test_size=0.2)
# Show length
print(len(input_tensor_train), len(target_tensor_train), len(input_tensor_val), len(target_tensor_val))
def convert(lang, tensor):
for t in tensor:
if t!=0:
print ("%d ----> %s" % (t, lang.index_word[t]))
print ("Input Language; index to word mapping")
convert(inp_lang, input_tensor_train[0])
print ()
print ("Target Language; index to word mapping")
convert(targ_lang, target_tensor_train[0])
BUFFER_SIZE = len(input_tensor_train)
BATCH_SIZE = 64
steps_per_epoch = len(input_tensor_train)//BATCH_SIZE
embedding_dim = 256
units = 1024
vocab_inp_size = len(inp_lang.word_index)+1
vocab_tar_size = len(targ_lang.word_index)+1
dataset = tf.data.Dataset.from_tensor_slices((input_tensor_train, target_tensor_train)).shuffle(BUFFER_SIZE)
dataset = dataset.batch(BATCH_SIZE, drop_remainder=True)
example_input_batch, example_target_batch = next(iter(dataset))
example_input_batch.shape, example_target_batch.shape
class Encoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, enc_units, batch_sz):
super(Encoder, self).__init__()
self.batch_sz = batch_sz
self.enc_units = enc_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = tf.keras.layers.GRU(self.enc_units,
return_sequences=True,
return_state=True,
recurrent_initializer='glorot_uniform')
def call(self, x, hidden):
x = self.embedding(x)
output, state = self.gru(x, initial_state = hidden)
return output, state
def initialize_hidden_state(self):
return tf.zeros((self.batch_sz, self.enc_units))
encoder = Encoder(vocab_inp_size, embedding_dim, units, BATCH_SIZE)
# sample input
sample_hidden = encoder.initialize_hidden_state()
sample_output, sample_hidden = encoder(example_input_batch, sample_hidden)
print ('Encoder output shape: (batch size, sequence length, units) {}'.format(sample_output.shape))
print ('Encoder Hidden state shape: (batch size, units) {}'.format(sample_hidden.shape))
class BahdanauAttention(tf.keras.Model):
def __init__(self, units):
super(BahdanauAttention, self).__init__()
self.W1 = tf.keras.layers.Dense(units)
self.W2 = tf.keras.layers.Dense(units)
self.V = tf.keras.layers.Dense(1)
def call(self, query, values):
# hidden shape == (batch_size, hidden size)
# hidden_with_time_axis shape == (batch_size, 1, hidden size)
# we are doing this to perform addition to calculate the score
hidden_with_time_axis = tf.expand_dims(query, 1)
# score shape == (batch_size, max_length, 1)
# we get 1 at the last axis because we are applying score to self.V
# the shape of the tensor before applying self.V is (batch_size, max_length, units)
score = self.V(tf.nn.tanh(
self.W1(values) + self.W2(hidden_with_time_axis)))
# attention_weights shape == (batch_size, max_length, 1)
attention_weights = tf.nn.softmax(score, axis=1)
# context_vector shape after sum == (batch_size, hidden_size)
context_vector = attention_weights * values
context_vector = tf.reduce_sum(context_vector, axis=1)
return context_vector, attention_weights
attention_layer = BahdanauAttention(10)
attention_result, attention_weights = attention_layer(sample_hidden, sample_output)
print("Attention result shape: (batch size, units) {}".format(attention_result.shape))
print("Attention weights shape: (batch_size, sequence_length, 1) {}".format(attention_weights.shape))
class Decoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, dec_units, batch_sz):
super(Decoder, self).__init__()
self.batch_sz = batch_sz
self.dec_units = dec_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = tf.keras.layers.GRU(self.dec_units,
return_sequences=True,
return_state=True,
recurrent_initializer='glorot_uniform')
self.fc = tf.keras.layers.Dense(vocab_size)
# used for attention
self.attention = BahdanauAttention(self.dec_units)
def call(self, x, hidden, enc_output):
# enc_output shape == (batch_size, max_length, hidden_size)
context_vector, attention_weights = self.attention(hidden, enc_output)
# x shape after passing through embedding == (batch_size, 1, embedding_dim)
x = self.embedding(x)
# x shape after concatenation == (batch_size, 1, embedding_dim + hidden_size)
x = tf.concat([tf.expand_dims(context_vector, 1), x], axis=-1)
# passing the concatenated vector to the GRU
output, state = self.gru(x)
# output shape == (batch_size * 1, hidden_size)
output = tf.reshape(output, (-1, output.shape[2]))
# output shape == (batch_size, vocab)
x = self.fc(output)
return x, state, attention_weights
decoder = Decoder(vocab_tar_size, embedding_dim, units, BATCH_SIZE)
sample_decoder_output, _, _ = decoder(tf.random.uniform((64, 1)),
sample_hidden, sample_output)
print ('Decoder output shape: (batch_size, vocab size) {}'.format(sample_decoder_output.shape))
optimizer = tf.keras.optimizers.Adam()
loss_object = tf.keras.losses.SparseCategoricalCrossentropy(
from_logits=True, reduction='none')
def loss_function(real, pred):
mask = tf.math.logical_not(tf.math.equal(real, 0))
loss_ = loss_object(real, pred)
mask = tf.cast(mask, dtype=loss_.dtype)
loss_ *= mask
return tf.reduce_mean(loss_)
checkpoint_dir = './training_checkpoints'
checkpoint_prefix = os.path.join(checkpoint_dir, "ckpt")
checkpoint = tf.train.Checkpoint(optimizer=optimizer,
encoder=encoder,
decoder=decoder)
@tf.function
def train_step(inp, targ, enc_hidden):
loss = 0
with tf.GradientTape() as tape:
enc_output, enc_hidden = encoder(inp, enc_hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word_index['<start>']] * BATCH_SIZE, 1)
# Teacher forcing - feeding the target as the next input
for t in range(1, targ.shape[1]):
# passing enc_output to the decoder
predictions, dec_hidden, _ = decoder(dec_input, dec_hidden, enc_output)
loss += loss_function(targ[:, t], predictions)
# using teacher forcing
dec_input = tf.expand_dims(targ[:, t], 1)
batch_loss = (loss / int(targ.shape[1]))
variables = encoder.trainable_variables + decoder.trainable_variables
gradients = tape.gradient(loss, variables)
optimizer.apply_gradients(zip(gradients, variables))
return batch_loss
EPOCHS = 10
for epoch in range(EPOCHS):
start = time.time()
enc_hidden = encoder.initialize_hidden_state()
total_loss = 0
for (batch, (inp, targ)) in enumerate(dataset.take(steps_per_epoch)):
batch_loss = train_step(inp, targ, enc_hidden)
total_loss += batch_loss
if batch % 100 == 0:
print('Epoch {} Batch {} Loss {:.4f}'.format(epoch + 1,
batch,
batch_loss.numpy()))
# saving (checkpoint) the model every 2 epochs
if (epoch + 1) % 2 == 0:
checkpoint.save(file_prefix = checkpoint_prefix)
print('Epoch {} Loss {:.4f}'.format(epoch + 1,
total_loss / steps_per_epoch))
print('Time taken for 1 epoch {} sec\n'.format(time.time() - start))
def evaluate(sentence):
attention_plot = np.zeros((max_length_targ, max_length_inp))
sentence = preprocess_sentence(sentence)
inputs = [inp_lang.word_index[i] for i in sentence.split(' ')]
inputs = tf.keras.preprocessing.sequence.pad_sequences([inputs],
maxlen=max_length_inp,
padding='post')
inputs = tf.convert_to_tensor(inputs)
result = ''
hidden = [tf.zeros((1, units))]
enc_out, enc_hidden = encoder(inputs, hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word_index['<start>']], 0)
for t in range(max_length_targ):
predictions, dec_hidden, attention_weights = decoder(dec_input,
dec_hidden,
enc_out)
# storing the attention weights to plot later on
attention_weights = tf.reshape(attention_weights, (-1, ))
attention_plot[t] = attention_weights.numpy()
predicted_id = tf.argmax(predictions[0]).numpy()
result += targ_lang.index_word[predicted_id] + ' '
if targ_lang.index_word[predicted_id] == '<end>':
return result, sentence, attention_plot
# the predicted ID is fed back into the model
dec_input = tf.expand_dims([predicted_id], 0)
return result, sentence, attention_plot
# function for plotting the attention weights
def plot_attention(attention, sentence, predicted_sentence):
fig = plt.figure(figsize=(10,10))
ax = fig.add_subplot(1, 1, 1)
ax.matshow(attention, cmap='viridis')
fontdict = {'fontsize': 14}
ax.set_xticklabels([''] + sentence, fontdict=fontdict, rotation=90)
ax.set_yticklabels([''] + predicted_sentence, fontdict=fontdict)
ax.xaxis.set_major_locator(ticker.MultipleLocator(1))
ax.yaxis.set_major_locator(ticker.MultipleLocator(1))
plt.show()
def translate(sentence):
result, sentence, attention_plot = evaluate(sentence)
print('Input: %s' % (sentence))
print('Predicted translation: {}'.format(result))
attention_plot = attention_plot[:len(result.split(' ')), :len(sentence.split(' '))]
plot_attention(attention_plot, sentence.split(' '), result.split(' '))
# restoring the latest checkpoint in checkpoint_dir
checkpoint.restore(tf.train.latest_checkpoint(checkpoint_dir))
translate(u'hace mucho frio aqui.')
translate(u'esta es mi vida.')
translate(u'¿todavia estan en casa?')
# wrong translation
translate(u'trata de averiguarlo.')
| 0.696887 | 0.904945 |
# Introduction
In this notebook, we demonstrate the steps needed to create an IoT Edge deployable module from the regression model created in the [turbofan regression](./turbofan_regression.ipynb) notebook. The steps we will follow are:
1. Reload experiment and model from the Azure Machine Learning service workspace
1. Create a scoring script
1. Create an environment YAML file
1. Create a container image using the model, scoring script and YAML file
1. Deploy the container image as a web service
1. Test the web service to make sure the container works as expected
1. Delete the web service
><font color=gray>Note: this notebook depends on the workspace, experiment and model created in the [turbofan regression](./turbofan_regression.ipynb) notebook.</font>
# Set up notebook
Please ensure that you are running this notebook under the Python 3.6 Kernel. The current kernel is show on the top of the notebook at the far right side of the file menu. If you are not running Python 3.6 you can change it in the file menu by clicking **Kernel->Change Kernel->Python 3.6**
```
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
%matplotlib inline
```
## Configure workspace
Create a workspace object from the existing workspace. `Workspace.from_config()` reads the file **aml_config/.azureml/config.json** and loads the details into an object named `ws`, which is used throughout the rest of the code in this notebook.
```
from azureml.core.workspace import Workspace
from azureml.core.experiment import Experiment
from azureml.core.model import Model
from azureml.train.automl.run import AutoMLRun
ws = Workspace.from_config(path='./aml_config')
```
## Load run, experiment and model
Use the model information that we persisted in the [turbofan regression](./turbofan_regression.ipynb) noebook to load our model.
```
import json
#name project folder and experiment
model_data = json.load(open('./aml_config/model_config.json'))
run_id = model_data['regressionRunId']
experiment_name = model_data['experimentName']
model_id = model_data['modelId']
experiment = Experiment(ws, experiment_name)
automl_run = AutoMLRun(experiment = experiment, run_id = run_id)
model = Model(ws, model_id)
```
# Create scoring script
The scoring script is the piece of code that runs inside the container and interacts with the model to return a prediction to the caller of web service or Azure IoT Edge module that is running the container. The scoring script is authored knowing the shape of the message that will be sent to the container. In our case, we have chosen to format the message as:
```json
[{
"DeviceId": 81,
"CycleTime": 140,
"OperationalSetting1": 0.0,
"OperationalSetting2": -0.0002,
"OperationalSetting3": 100.0,
"Sensor1": 518.67,
"Sensor2": 642.43,
"Sensor3": 1596.02,
"Sensor4": 1404.4,
"Sensor5": 14.62,
"Sensor6": 21.6,
"Sensor7": 559.76,
"Sensor8": 2388.19,
"Sensor9": 9082.16,
"Sensor10": 1.31,
"Sensor11": 47.6,
"Sensor12": 527.82,
"Sensor13": 2388.17,
"Sensor14": 8155.92,
"Sensor15": 8.3214,
"Sensor16": 0.03,
"Sensor17": 393.0,
"Sensor18": 2388.0,
"Sensor19": 100.0,
"Sensor20": 39.41,
"Sensor21": 23.5488
}]
```
><font color='gray'>See the [Azure IoT Edge ML whitepaper](https://aka.ms/IoTEdgeMLPaper) for details about how messages are formatted and sent to the classifier module.</font>
```
script_file_name = 'score.py'
%%writefile $script_file_name
import pickle
import json
import numpy as np
import pandas as pd
import azureml.train.automl
from sklearn.externals import joblib
from azureml.core.model import Model
def init():
global model
model_path = Model.get_model_path(model_name = '<<modelname>>')
# deserialize the model file back into a sklearn model
model = joblib.load(model_path)
def unpack_message(raw_data):
message_data = json.loads(raw_data)
# convert single message to list
if type(message_data) is dict:
message_data = [message_data]
return message_data
def extract_features(message_data):
X_data = []
sensor_names = ['Sensor'+str(i) for i in range(1,22)]
for message in message_data:
# select sensor data from the message dictionary
feature_dict = {k: message[k] for k in (sensor_names)}
X_data.append(feature_dict)
X_df = pd.DataFrame(X_data)
return np.array(X_df[sensor_names].values)
def append_predict_data(message_data, y_hat):
message_df = pd.DataFrame(message_data)
message_df['PredictedRul'] = y_hat
return message_df.to_dict('records')
def log_for_debug(log_message, log_data):
print("*****%s:" % log_message)
print(log_data)
print("******")
def run(raw_data):
log_for_debug("raw_data", raw_data)
message_data = unpack_message(raw_data)
log_for_debug("message_data", message_data)
X_data = extract_features(message_data)
log_for_debug("X_data", X_data)
# make prediction
y_hat = model.predict(X_data)
response_data = append_predict_data(message_data, y_hat)
return response_data
```
### Update the scoring script with the actual model ID
```
# Substitute the actual model id in the script file.
with open(script_file_name, 'r') as cefr:
content = cefr.read()
with open(script_file_name, 'w') as cefw:
cefw.write(content.replace('<<modelname>>', model.name))
```
## Create YAML file for the environment
The YAML file provides the information about the dependencies for the model we will deploy.
### Get azureml versions
First we will use the run to retrieve the version of the azureml packages used to train the model.
>Warnings about the version of the SDK not matching with the training version are expected
```
best_run, fitted_model = automl_run.get_output()
iteration = int(best_run.get_properties()['iteration'])
dependencies = automl_run.get_run_sdk_dependencies(iteration = iteration)
for p in ['azureml-train-automl', 'azureml-sdk', 'azureml-core']:
print('{}\t{}'.format(p, dependencies[p]))
```
### Write YAML file
Write the initial YAML file to disk and update the dependencies for azureml to match with the training versions. This is not strictly needed in this notebook because the model likely has been generated using the current SDK version. However, we include this for completeness for the case when an experiment was trained using a previous SDK version.
```
import azureml.core
from azureml.core.conda_dependencies import CondaDependencies
myenv = CondaDependencies.create(conda_packages=['numpy','scikit-learn','pandas'], pip_packages=['azureml-sdk[automl]'])
conda_env_file_name = 'myenv.yml'
myenv.save_to_file('.', conda_env_file_name)
# Substitute the actual version number in the environment file.
with open(conda_env_file_name, 'r') as cefr:
content = cefr.read()
with open(conda_env_file_name, 'w') as cefw:
cefw.write(content.replace(azureml.core.VERSION, dependencies['azureml-sdk']))
```
## Create a container image
Use the scoring script and the YAML file to create a container image in the workspace. The image will take several minutes to create.
```
from azureml.core.image import Image, ContainerImage
image_config = ContainerImage.image_configuration(runtime= "python",
execution_script = script_file_name,
conda_file = conda_env_file_name,
tags = {'area': "digits", 'type': "automl_classification"},
description = "Image for Edge ML samples")
image = Image.create(name = "edgemlsample",
# this is the model object
models = [model],
image_config = image_config,
workspace = ws)
image.wait_for_creation(show_output = True)
if image.creation_state == 'Failed':
print("Image build log at: " + image.image_build_log_uri)
```
## Deploy image as a web service on Azure Container Instance
Deploy the image we just created as web service on Azure Container Instance (ACI). We will use this web service to test that our model/container performs as expected.
```
from azureml.core.webservice import AciWebservice
from azureml.core.webservice import Webservice
aci_service_name = 'edge-ml-rul-01'
aci_config = AciWebservice.deploy_configuration(cpu_cores = 1,
memory_gb = 1,
tags = {'area': "digits", 'type': "automl_RUL"},
description = 'test service for Edge ML RUL')
print ("Deploying service: %s" % aci_service_name)
aci_service = Webservice.deploy_from_image(deployment_config = aci_config,
image = image,
name = aci_service_name,
workspace = ws)
aci_service.wait_for_deployment(True)
print ("Service state: %s" % aci_service.state)
```
## Load test data
To save a couple of steps at this point, we serialized the test data that we loaded in the [turbofan regression](./turbofan_regression.ipynb) notebook. Here we deserialize that data to use it to test the web service.
```
import pandas as pd
from sklearn.externals import joblib
import numpy
test_df = pd.read_csv("data/WebServiceTest.csv")
test_df.head(5)
```
## Predict one message at a time
Once the container/model is deployed to and Azure IoT Edge device it will receive messages one at a time. Send a few messages in that mode to make sure everything is working.
```
import json
import pandas as pd
# reformat data as list of messages
X_message = test_df.head(5).to_dict('record')
result_list = []
for row in X_message:
row_data = json.dumps(row)
row_result = aci_service.run(input_data=row_data)
result_list.append(row_result[0])
result_df = pd.DataFrame(result_list)
residuals = result_df['RUL'] - result_df['PredictedRul']
result_df['Residual'] = residuals
result_df[['CycleTime','RUL','PredictedRul','Residual']]
```
## Predict entire set
To make sure the model as a whole is working as expected, we send the test set in bulk to the model, save the predictions, and calculate the residual.
```
import json
import pandas as pd
X_messages = test_df.to_dict('record')
raw_data = json.dumps(X_messages)
result_list = aci_service.run(input_data=raw_data)
result_df = pd.DataFrame(result_list)
residuals = result_df['RUL'] - result_df['PredictedRul']
result_df['Residual'] = residuals
y_test = result_df['RUL']
y_pred = result_df['PredictedRul']
```
## Plot actuals vs. predicted
To validate the shape of the model, plot the actual RUL against the predicted RUL for each cycle and device.
```
from sklearn.metrics import mean_squared_error, r2_score
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
fig, ax = plt.subplots()
fig.set_size_inches(8, 4)
font_size = 12
g = sns.regplot(y='PredictedRul', x='RUL', data=result_df, fit_reg=False, ax=ax)
lim_set = g.set(ylim=(0, 500), xlim=(0, 500))
plot = g.axes.plot([0, 500], [0, 500], c=".3", ls="--");
rmse = ax.text(16,450,'RMSE = {0:.2f}'.format(numpy.sqrt(mean_squared_error(y_test, y_pred))), fontsize = font_size)
r2 = ax.text(16,425,'R2 Score = {0:.2f}'.format(r2_score(y_test, y_pred)), fontsize = font_size)
xlabel = ax.set_xlabel('Actual RUL', size=font_size)
ylabel = ax.set_ylabel('Predicted RUL', size=font_size)
```
## Delete web service
Now that we are confident that our container and model are working well, delete the web service.
```
from azureml.core.webservice import Webservice
aci_service = Webservice(ws, 'edge-ml-rul-01')
aci_service.delete()
```
|
github_jupyter
|
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
%matplotlib inline
from azureml.core.workspace import Workspace
from azureml.core.experiment import Experiment
from azureml.core.model import Model
from azureml.train.automl.run import AutoMLRun
ws = Workspace.from_config(path='./aml_config')
import json
#name project folder and experiment
model_data = json.load(open('./aml_config/model_config.json'))
run_id = model_data['regressionRunId']
experiment_name = model_data['experimentName']
model_id = model_data['modelId']
experiment = Experiment(ws, experiment_name)
automl_run = AutoMLRun(experiment = experiment, run_id = run_id)
model = Model(ws, model_id)
[{
"DeviceId": 81,
"CycleTime": 140,
"OperationalSetting1": 0.0,
"OperationalSetting2": -0.0002,
"OperationalSetting3": 100.0,
"Sensor1": 518.67,
"Sensor2": 642.43,
"Sensor3": 1596.02,
"Sensor4": 1404.4,
"Sensor5": 14.62,
"Sensor6": 21.6,
"Sensor7": 559.76,
"Sensor8": 2388.19,
"Sensor9": 9082.16,
"Sensor10": 1.31,
"Sensor11": 47.6,
"Sensor12": 527.82,
"Sensor13": 2388.17,
"Sensor14": 8155.92,
"Sensor15": 8.3214,
"Sensor16": 0.03,
"Sensor17": 393.0,
"Sensor18": 2388.0,
"Sensor19": 100.0,
"Sensor20": 39.41,
"Sensor21": 23.5488
}]
script_file_name = 'score.py'
%%writefile $script_file_name
import pickle
import json
import numpy as np
import pandas as pd
import azureml.train.automl
from sklearn.externals import joblib
from azureml.core.model import Model
def init():
global model
model_path = Model.get_model_path(model_name = '<<modelname>>')
# deserialize the model file back into a sklearn model
model = joblib.load(model_path)
def unpack_message(raw_data):
message_data = json.loads(raw_data)
# convert single message to list
if type(message_data) is dict:
message_data = [message_data]
return message_data
def extract_features(message_data):
X_data = []
sensor_names = ['Sensor'+str(i) for i in range(1,22)]
for message in message_data:
# select sensor data from the message dictionary
feature_dict = {k: message[k] for k in (sensor_names)}
X_data.append(feature_dict)
X_df = pd.DataFrame(X_data)
return np.array(X_df[sensor_names].values)
def append_predict_data(message_data, y_hat):
message_df = pd.DataFrame(message_data)
message_df['PredictedRul'] = y_hat
return message_df.to_dict('records')
def log_for_debug(log_message, log_data):
print("*****%s:" % log_message)
print(log_data)
print("******")
def run(raw_data):
log_for_debug("raw_data", raw_data)
message_data = unpack_message(raw_data)
log_for_debug("message_data", message_data)
X_data = extract_features(message_data)
log_for_debug("X_data", X_data)
# make prediction
y_hat = model.predict(X_data)
response_data = append_predict_data(message_data, y_hat)
return response_data
# Substitute the actual model id in the script file.
with open(script_file_name, 'r') as cefr:
content = cefr.read()
with open(script_file_name, 'w') as cefw:
cefw.write(content.replace('<<modelname>>', model.name))
best_run, fitted_model = automl_run.get_output()
iteration = int(best_run.get_properties()['iteration'])
dependencies = automl_run.get_run_sdk_dependencies(iteration = iteration)
for p in ['azureml-train-automl', 'azureml-sdk', 'azureml-core']:
print('{}\t{}'.format(p, dependencies[p]))
import azureml.core
from azureml.core.conda_dependencies import CondaDependencies
myenv = CondaDependencies.create(conda_packages=['numpy','scikit-learn','pandas'], pip_packages=['azureml-sdk[automl]'])
conda_env_file_name = 'myenv.yml'
myenv.save_to_file('.', conda_env_file_name)
# Substitute the actual version number in the environment file.
with open(conda_env_file_name, 'r') as cefr:
content = cefr.read()
with open(conda_env_file_name, 'w') as cefw:
cefw.write(content.replace(azureml.core.VERSION, dependencies['azureml-sdk']))
from azureml.core.image import Image, ContainerImage
image_config = ContainerImage.image_configuration(runtime= "python",
execution_script = script_file_name,
conda_file = conda_env_file_name,
tags = {'area': "digits", 'type': "automl_classification"},
description = "Image for Edge ML samples")
image = Image.create(name = "edgemlsample",
# this is the model object
models = [model],
image_config = image_config,
workspace = ws)
image.wait_for_creation(show_output = True)
if image.creation_state == 'Failed':
print("Image build log at: " + image.image_build_log_uri)
from azureml.core.webservice import AciWebservice
from azureml.core.webservice import Webservice
aci_service_name = 'edge-ml-rul-01'
aci_config = AciWebservice.deploy_configuration(cpu_cores = 1,
memory_gb = 1,
tags = {'area': "digits", 'type': "automl_RUL"},
description = 'test service for Edge ML RUL')
print ("Deploying service: %s" % aci_service_name)
aci_service = Webservice.deploy_from_image(deployment_config = aci_config,
image = image,
name = aci_service_name,
workspace = ws)
aci_service.wait_for_deployment(True)
print ("Service state: %s" % aci_service.state)
import pandas as pd
from sklearn.externals import joblib
import numpy
test_df = pd.read_csv("data/WebServiceTest.csv")
test_df.head(5)
import json
import pandas as pd
# reformat data as list of messages
X_message = test_df.head(5).to_dict('record')
result_list = []
for row in X_message:
row_data = json.dumps(row)
row_result = aci_service.run(input_data=row_data)
result_list.append(row_result[0])
result_df = pd.DataFrame(result_list)
residuals = result_df['RUL'] - result_df['PredictedRul']
result_df['Residual'] = residuals
result_df[['CycleTime','RUL','PredictedRul','Residual']]
import json
import pandas as pd
X_messages = test_df.to_dict('record')
raw_data = json.dumps(X_messages)
result_list = aci_service.run(input_data=raw_data)
result_df = pd.DataFrame(result_list)
residuals = result_df['RUL'] - result_df['PredictedRul']
result_df['Residual'] = residuals
y_test = result_df['RUL']
y_pred = result_df['PredictedRul']
from sklearn.metrics import mean_squared_error, r2_score
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
fig, ax = plt.subplots()
fig.set_size_inches(8, 4)
font_size = 12
g = sns.regplot(y='PredictedRul', x='RUL', data=result_df, fit_reg=False, ax=ax)
lim_set = g.set(ylim=(0, 500), xlim=(0, 500))
plot = g.axes.plot([0, 500], [0, 500], c=".3", ls="--");
rmse = ax.text(16,450,'RMSE = {0:.2f}'.format(numpy.sqrt(mean_squared_error(y_test, y_pred))), fontsize = font_size)
r2 = ax.text(16,425,'R2 Score = {0:.2f}'.format(r2_score(y_test, y_pred)), fontsize = font_size)
xlabel = ax.set_xlabel('Actual RUL', size=font_size)
ylabel = ax.set_ylabel('Predicted RUL', size=font_size)
from azureml.core.webservice import Webservice
aci_service = Webservice(ws, 'edge-ml-rul-01')
aci_service.delete()
| 0.374791 | 0.942401 |
```
import requests as r
%matplotlib inline
import pandas as pd
from nltk.corpus import stopwords
#we don't need these. I left them here just in case.
# appID = '1844787218898682'
# appSecret = 'b3e7536d6dd029c773441d5271318dec'
#this is where you have to go to get a token to test with. You need to go there to get a new access_token every time you run the script.
#https://developers.facebook.com/tools/explorer/145634995501895/?method=GET&path=me%2Ffeed&version=v2.10
access_token = 'EAACEdEose0cBAGHWZAec37neJdulqMocs6LZCMQUrn8H0Er2nNtSFLBeFmtR4DCiZC9gVYSTXeKykqlbiOU9oUhEZAnl5vJ55aYOLYcOXOPbveW5sc9VdPh9C0TVecAOZBV0L7wdQbdZB9mjI9JtEQJEuIEtYrWycrLw5CpBMw2u29VKxyVQhLhZCsz5OvqfloZD'
allData = []
firstURL = 'https://graph.facebook.com/v2.10/me/posts?access_token={}'.format(access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
while 'paging' in postData:
nextURL = postData['paging']['next']
response = r.get(nextURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
df = pd.DataFrame(allData)
df.created_time = pd.to_datetime(df.created_time)
df = df[df.created_time > datetime.datetime(2011,6,1)].copy()
text = df[df.message == df.message].message.str.cat(sep=' ')
text2 = text.replace('\n\n',' ').replace('\n',' ').replace(u"\'","").lower()
test = pd.Series(text2.split(' '))
urls = test[test.str.contains('http')|test.str.contains('www')].values
test = test[~(test.str.contains('http')|test.str.contains('www'))].copy()
text2 = test.str.cat(sep=' ')
import string
for x in list(string.punctuation):
text2 = text2.replace(x,' ')
words_list = text2.split(' ')
words_list = pd.Series(words_list)
words_list = words_list[words_list != ''].copy()
stop = stopwords.words('english')
final_words_list = words_list[~words_list.isin(stop)]
#top words used in my Facebook posts
final_words_list.value_counts()[:20]
from textblob import TextBlob
dfNoNan = df[df.message == df.message].copy()
dfNoNan['polarity'] = dfNoNan['message'].map(lambda x: TextBlob(x).sentiment.polarity)
dfNoNan['subjectivity'] = dfNoNan['message'].map(lambda x: TextBlob(x).sentiment.subjectivity)
import math
result = {}
for x in range(math.ceil(len(dfNoNan.id) / 50)):
newIDs = dfNoNan.id[50*x:50*(x+1)]
stringIDs = ','.join(newIDs)
firstURL = 'https://graph.facebook.com/v2.10?ids={}&fields=shares.limit(5000).summary(true),likes.limit(5000).summary(true),comments.limit(5000).summary(true)&access_token={}'.format(stringIDs,access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
result.update(postData)
print(len(result))
def getLikesName(cell):
total_likes_name = len(result[cell]['likes']['data'])
return total_likes_name
def getLikesCount(cell):
total_likes_count = result[cell]['likes']['summary']['total_count']
return total_likes_count
def getCommentsName(cell):
total_comments_name = len(result[cell]['comments']['data'])
return total_comments_name
def getCommentsCount(cell):
total_comments_count = result[cell]['comments']['summary']['total_count']
return total_comments_count
dfNoNan['likesName'] = dfNoNan.id.map(getLikesName)
dfNoNan['likesCount'] = dfNoNan.id.map(getLikesCount)
dfNoNan['commentsName'] = dfNoNan.id.map(getCommentsName)
dfNoNan['commentsCount'] = dfNoNan.id.map(getCommentsCount)
#total number of posts on facebook (this counts shares and posts)
df['id'].count()
#total number of times posts have been liked
dfNoNan.likesCount.sum()
likes_people = []
comments_people = []
for postID in dfNoNan.id:
likes_people = likes_people + [x['name'] for x in result[postID]['likes']['data']]
comments_people = comments_people + [x['from']['name'] for x in result[postID]['comments']['data']]
likes_people = pd.Series(likes_people)
comments_people = pd.Series(comments_people)
#top people based on comments on my Facebook Posts
comments_people.value_counts()[:20]
#top people based on likes on my Facebook Posts
likes_people.value_counts()[:20]
#top tweets based on comments
dfNoNan.sort_values('commentsCount',ascending=False)[:10][['created_time','message','story','commentsCount']]
#top tweets based on likes
dfNoNan.sort_values('likesCount',ascending=False)[:10][['created_time','message','story','likesCount']]
import datetime
df.created_time = pd.to_datetime(df.created_time)
timedf = df.copy()
timedf.index = df.created_time
test = timedf.groupby(timedf.index.year.astype(str) + timedf.index.week.astype(str)).count()
def fixIndex(cell):
if len(cell) == 5:
return cell[:4] + '0' + cell[4]
else:
return cell
test.index = test.index.map(fixIndex)
new_week_index = []
#eventually pull these dates from latest tweet, go back until earliest tweet
for x in range(2011,2018):
for y in range(0,53):
new_week_index.append(str(x) + str(y))
new_week_index = pd.Series(new_week_index).map(fixIndex)
test = test.reindex(new_week_index, fill_value=0)
#facebook activity over time (grouped by week)
test['id'].plot()
test2 = timedf.groupby(timedf.index.dayofweek).count()
#Monday is 0. Sunday is 6.
#Facebook posts by day of week
test2['id'].plot()
#this is because I am in a time zone that is -7 GMT
#this will need to be changed based on user time zone
test3 = timedf.groupby(timedf.index.hour - 7).count()
#0 = midnight. 12 = noon.
#facebook posts by time of day
test3['id'].plot()
test4 = timedf.groupby(timedf.index.month).count()
#facebook posts by month
test4['id'].plot()
allData = []
firstURL = 'https://graph.facebook.com/v2.10/10153451802413555/likes?access_token={}'.format(access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
while 'paging' in postData:
nextURL = postData['paging']['next']
response = r.get(nextURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
pages = pd.DataFrame(allData)
#get the feed
allData = []
firstURL = 'https://graph.facebook.com/v2.10/10153451802413555/feed?access_token={}'.format(access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
while 'paging' in postData:
nextURL = postData['paging']['next']
response = r.get(nextURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
feed = pd.DataFrame(allData)
import math
result = {}
for x in range(math.ceil(len(feed.id) / 50)):
newIDs = feed.id[50*x:50*(x+1)]
stringIDs = ','.join(newIDs)
firstURL = 'https://graph.facebook.com/v2.10?ids={}&fields=shares.limit(5000).summary(true),likes.limit(5000).summary(true),comments.limit(5000).summary(true)&access_token={}'.format(stringIDs,access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
result.update(postData)
print(len(result))
#this is your Facebook feed (not just your posts, but everything that shows up on your wall)
#this can go back into your other script and you can look at all of the same things as just the posts script
#result
#get og.likes (only returns 3 likes, they are all associated with GoFundMe. I don't know why.)
allData = []
firstURL = 'https://graph.facebook.com/v2.10/10153451802413555/og.likes?since=1167609600&access_token={}'.format(access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
#scroll down in activity viewer to get all likes and comments
#use copy(document.body.innerHTML); to copy text
#paste it somewhere. Save it.
#now analyze it.
with open('C:/Users/bodil/Desktop/FacebookPostsLikes.html','r',encoding='utf-8') as file:
data = file.read()
from bs4 import BeautifulSoup as BS
soup = BS(data,"lxml")
soup2 = soup.find_all('div',class_='fbTimelineLogBody')
profiles = soup2[0].find_all('a',class_="profileLink")
people = soup2[0].find_all('td',class_='_5ep5')
posts = soup2[0].find_all('div',class_='fsm')
people2 = []
for x in people:
people2.append(x.text)
def parse_comments(cell):
flag = False
if 'his own comment' in cell:
return 'his own comment'
if 'comment' in cell and 'commented' not in cell:
return 'comment'
elif 'post' in cell:
return 'post'
elif 'life event' in cell:
return 'life event'
elif 'link' in cell:
return 'link'
elif 'album' in cell:
return 'album'
elif 'photo' in cell:
return 'photo'
elif 'live video' in cell:
return 'live video'
elif 'video' in cell:
return 'video'
elif 'campaign' in cell:
return 'campaign'
elif 'status' in cell:
return 'status'
elif 'bio' in cell:
return 'bio'
elif 'timeline' in cell:
return 'timeline'
elif 'friend request' in cell:
return 'friend request'
elif 'profile picture' in cell:
return 'profile picture'
elif 'an event' in cell:
return 'an event'
elif 'a list' in cell:
return 'a list'
elif 'an entry' in cell:
return 'an entry'
elif 'a note' in cell:
return 'a note'
elif 'Quotations' in cell:
return 'quotations'
elif 'phone number' in cell:
return 'phone number'
elif 'Website' in cell:
return 'website'
elif 'memory' in cell:
return 'memory'
elif 'poll' in cell:
return 'poll'
elif 'hometown' in cell:
return 'hometown'
elif 'Page' in cell:
return 'page'
else:
return None
people2 = pd.DataFrame(people2)
# people2[~(people2[0].map(parse_comments)==people2[0].map(parse_comments))]
people2['media'] = people2[0].map(parse_comments)
#top media that you interact with
people2.media.value_counts()[:10]
people2.rename(columns={0:'event_text'},inplace=True)
def parse_action(cell):
if 'liked' in cell:
return 'liked'
elif 'likes' in cell:
return 'likes'
elif 'reacted to' in cell:
return 'reacted to'
elif 'is going to' in cell:
return 'is going to'
elif 'was mentioned in a' in cell:
return 'was mentioned in a'
elif 'commented on' in cell:
return 'commented on'
elif 'posted in' in cell:
return 'posted in'
elif 'became friends with' in cell:
return 'became friends with'
elif 'replied to' in cell:
return 'replied to'
elif 'added' in cell:
return 'added'
elif 'shared a post to your' in cell:
return 'shared a post to your'
elif 'was tagged in' in cell:
return 'was tagged in'
elif 'shared' in cell:
return 'shared'
elif 'wrote on your' in cell:
return 'wrote on your'
elif 'wrote on' in cell:
return 'wrote on'
elif 'updated' in cell:
return 'updated'
elif 'sent' in cell:
return 'sent'
elif 'changed' in cell:
return 'changed'
elif 'posted a video to' in cell:
return 'posted a video to'
elif 'posted something via' in cell:
return 'posted something via'
elif 'voted for' in cell:
return 'voted for'
elif 'was feeling' in cell:
return 'was feeling'
elif 'reviewed' in cell:
return 'reviewed'
elif 'created' in cell:
return 'created'
elif 'was tagged at' in cell:
return 'was tagged at'
elif 'voted on' in cell:
return 'voted on'
elif 'was with' in cell:
return 'was with'
elif 'is now using Facebook in' in cell:
return 'is now using Facebook in'
elif 'is interested in' in cell:
return 'is interested in'
elif 'was' in cell:
return 'was'
# people2[~(people2.event_text.map(parse_action)==people2.event_text.map(parse_action))]
people2['event'] = people2.event_text.map(parse_action)
#top interactions on Facebook
people2.event.value_counts()[:10]
people2['date'] = people2.event_text.map(lambda x: x.split('.')[-1])
for x in people2.index:
if people2.loc[x]['event_text'] != people2.loc[x]['event_text']:
people2.loc[x,'name'] = None
else:
test = people2.loc[x]['event_text']
if people2.loc[x,'event'] == 'became friends with':
person = test[test.find(people2.loc[x]['event']) + len(people2.loc[x]['event']) + 1:test.find('.')]
people2.loc[x,'name'] = person
elif ' own ' in people2.loc[x,'event_text'] or ' his ' in people2.loc[x,'event_text'] or ' her ' in people2.loc[x,'event_text']:
people2.loc[x,'name'] = None
elif people2.loc[x,'event'] == 'added':
people2.loc[x,'name'] = None
elif people2.loc[x,'media'] == None:
people2.loc[x,'name'] = None
else:
person = test[test.find(people2.loc[x]['event']) + len(people2.loc[x]['event']) + 1:test.find(people2.loc[x]['media']) - 3]
people2.loc[x,'name'] = person
#people you interact with the most
people2[people2.name != ''].name.value_counts()[:20]
#things you interact with the most
people2.media.value_counts()[:10]
posts2 = []
for x in posts:
posts2.append(x.text)
text = ' '.join(posts2)
text2 = text.replace('\n\n',' ').replace('\n',' ').replace(u"\'","").lower()
test = pd.Series(text2.split(' '))
urls = test[test.str.contains('http')|test.str.contains('www')].values
test = test[~(test.str.contains('http')|test.str.contains('www')|test.str.contains('.com'))].copy()
text2 = test.str.cat(sep=' ')
import string
for x in list(string.punctuation):
text2 = text2.replace(x,' ')
words_list = text2.split(' ')
words_list = pd.Series(words_list)
words_list = words_list[words_list != ''].copy()
stop = stopwords.words('english')
final_words_list = words_list[~words_list.isin(stop)]
#words in Facebook interactions you have had
final_words_list.value_counts()
#look at likes and comments over time for a single post (top liked and commented posts--to get a sense for timeline of liking and posting)
#when do I like, comment, or post things? Any patterns?
#look at URLs I have posted about (top URLs...?)
#look at hashtags I have used (how am I treating #aslkdflksd--I need to pull them out)
```
|
github_jupyter
|
import requests as r
%matplotlib inline
import pandas as pd
from nltk.corpus import stopwords
#we don't need these. I left them here just in case.
# appID = '1844787218898682'
# appSecret = 'b3e7536d6dd029c773441d5271318dec'
#this is where you have to go to get a token to test with. You need to go there to get a new access_token every time you run the script.
#https://developers.facebook.com/tools/explorer/145634995501895/?method=GET&path=me%2Ffeed&version=v2.10
access_token = 'EAACEdEose0cBAGHWZAec37neJdulqMocs6LZCMQUrn8H0Er2nNtSFLBeFmtR4DCiZC9gVYSTXeKykqlbiOU9oUhEZAnl5vJ55aYOLYcOXOPbveW5sc9VdPh9C0TVecAOZBV0L7wdQbdZB9mjI9JtEQJEuIEtYrWycrLw5CpBMw2u29VKxyVQhLhZCsz5OvqfloZD'
allData = []
firstURL = 'https://graph.facebook.com/v2.10/me/posts?access_token={}'.format(access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
while 'paging' in postData:
nextURL = postData['paging']['next']
response = r.get(nextURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
df = pd.DataFrame(allData)
df.created_time = pd.to_datetime(df.created_time)
df = df[df.created_time > datetime.datetime(2011,6,1)].copy()
text = df[df.message == df.message].message.str.cat(sep=' ')
text2 = text.replace('\n\n',' ').replace('\n',' ').replace(u"\'","").lower()
test = pd.Series(text2.split(' '))
urls = test[test.str.contains('http')|test.str.contains('www')].values
test = test[~(test.str.contains('http')|test.str.contains('www'))].copy()
text2 = test.str.cat(sep=' ')
import string
for x in list(string.punctuation):
text2 = text2.replace(x,' ')
words_list = text2.split(' ')
words_list = pd.Series(words_list)
words_list = words_list[words_list != ''].copy()
stop = stopwords.words('english')
final_words_list = words_list[~words_list.isin(stop)]
#top words used in my Facebook posts
final_words_list.value_counts()[:20]
from textblob import TextBlob
dfNoNan = df[df.message == df.message].copy()
dfNoNan['polarity'] = dfNoNan['message'].map(lambda x: TextBlob(x).sentiment.polarity)
dfNoNan['subjectivity'] = dfNoNan['message'].map(lambda x: TextBlob(x).sentiment.subjectivity)
import math
result = {}
for x in range(math.ceil(len(dfNoNan.id) / 50)):
newIDs = dfNoNan.id[50*x:50*(x+1)]
stringIDs = ','.join(newIDs)
firstURL = 'https://graph.facebook.com/v2.10?ids={}&fields=shares.limit(5000).summary(true),likes.limit(5000).summary(true),comments.limit(5000).summary(true)&access_token={}'.format(stringIDs,access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
result.update(postData)
print(len(result))
def getLikesName(cell):
total_likes_name = len(result[cell]['likes']['data'])
return total_likes_name
def getLikesCount(cell):
total_likes_count = result[cell]['likes']['summary']['total_count']
return total_likes_count
def getCommentsName(cell):
total_comments_name = len(result[cell]['comments']['data'])
return total_comments_name
def getCommentsCount(cell):
total_comments_count = result[cell]['comments']['summary']['total_count']
return total_comments_count
dfNoNan['likesName'] = dfNoNan.id.map(getLikesName)
dfNoNan['likesCount'] = dfNoNan.id.map(getLikesCount)
dfNoNan['commentsName'] = dfNoNan.id.map(getCommentsName)
dfNoNan['commentsCount'] = dfNoNan.id.map(getCommentsCount)
#total number of posts on facebook (this counts shares and posts)
df['id'].count()
#total number of times posts have been liked
dfNoNan.likesCount.sum()
likes_people = []
comments_people = []
for postID in dfNoNan.id:
likes_people = likes_people + [x['name'] for x in result[postID]['likes']['data']]
comments_people = comments_people + [x['from']['name'] for x in result[postID]['comments']['data']]
likes_people = pd.Series(likes_people)
comments_people = pd.Series(comments_people)
#top people based on comments on my Facebook Posts
comments_people.value_counts()[:20]
#top people based on likes on my Facebook Posts
likes_people.value_counts()[:20]
#top tweets based on comments
dfNoNan.sort_values('commentsCount',ascending=False)[:10][['created_time','message','story','commentsCount']]
#top tweets based on likes
dfNoNan.sort_values('likesCount',ascending=False)[:10][['created_time','message','story','likesCount']]
import datetime
df.created_time = pd.to_datetime(df.created_time)
timedf = df.copy()
timedf.index = df.created_time
test = timedf.groupby(timedf.index.year.astype(str) + timedf.index.week.astype(str)).count()
def fixIndex(cell):
if len(cell) == 5:
return cell[:4] + '0' + cell[4]
else:
return cell
test.index = test.index.map(fixIndex)
new_week_index = []
#eventually pull these dates from latest tweet, go back until earliest tweet
for x in range(2011,2018):
for y in range(0,53):
new_week_index.append(str(x) + str(y))
new_week_index = pd.Series(new_week_index).map(fixIndex)
test = test.reindex(new_week_index, fill_value=0)
#facebook activity over time (grouped by week)
test['id'].plot()
test2 = timedf.groupby(timedf.index.dayofweek).count()
#Monday is 0. Sunday is 6.
#Facebook posts by day of week
test2['id'].plot()
#this is because I am in a time zone that is -7 GMT
#this will need to be changed based on user time zone
test3 = timedf.groupby(timedf.index.hour - 7).count()
#0 = midnight. 12 = noon.
#facebook posts by time of day
test3['id'].plot()
test4 = timedf.groupby(timedf.index.month).count()
#facebook posts by month
test4['id'].plot()
allData = []
firstURL = 'https://graph.facebook.com/v2.10/10153451802413555/likes?access_token={}'.format(access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
while 'paging' in postData:
nextURL = postData['paging']['next']
response = r.get(nextURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
pages = pd.DataFrame(allData)
#get the feed
allData = []
firstURL = 'https://graph.facebook.com/v2.10/10153451802413555/feed?access_token={}'.format(access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
while 'paging' in postData:
nextURL = postData['paging']['next']
response = r.get(nextURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
feed = pd.DataFrame(allData)
import math
result = {}
for x in range(math.ceil(len(feed.id) / 50)):
newIDs = feed.id[50*x:50*(x+1)]
stringIDs = ','.join(newIDs)
firstURL = 'https://graph.facebook.com/v2.10?ids={}&fields=shares.limit(5000).summary(true),likes.limit(5000).summary(true),comments.limit(5000).summary(true)&access_token={}'.format(stringIDs,access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
result.update(postData)
print(len(result))
#this is your Facebook feed (not just your posts, but everything that shows up on your wall)
#this can go back into your other script and you can look at all of the same things as just the posts script
#result
#get og.likes (only returns 3 likes, they are all associated with GoFundMe. I don't know why.)
allData = []
firstURL = 'https://graph.facebook.com/v2.10/10153451802413555/og.likes?since=1167609600&access_token={}'.format(access_token)
response = r.get(firstURL)
print(response)
postData = response.json()
allData = allData + postData['data']
print(len(allData))
#scroll down in activity viewer to get all likes and comments
#use copy(document.body.innerHTML); to copy text
#paste it somewhere. Save it.
#now analyze it.
with open('C:/Users/bodil/Desktop/FacebookPostsLikes.html','r',encoding='utf-8') as file:
data = file.read()
from bs4 import BeautifulSoup as BS
soup = BS(data,"lxml")
soup2 = soup.find_all('div',class_='fbTimelineLogBody')
profiles = soup2[0].find_all('a',class_="profileLink")
people = soup2[0].find_all('td',class_='_5ep5')
posts = soup2[0].find_all('div',class_='fsm')
people2 = []
for x in people:
people2.append(x.text)
def parse_comments(cell):
flag = False
if 'his own comment' in cell:
return 'his own comment'
if 'comment' in cell and 'commented' not in cell:
return 'comment'
elif 'post' in cell:
return 'post'
elif 'life event' in cell:
return 'life event'
elif 'link' in cell:
return 'link'
elif 'album' in cell:
return 'album'
elif 'photo' in cell:
return 'photo'
elif 'live video' in cell:
return 'live video'
elif 'video' in cell:
return 'video'
elif 'campaign' in cell:
return 'campaign'
elif 'status' in cell:
return 'status'
elif 'bio' in cell:
return 'bio'
elif 'timeline' in cell:
return 'timeline'
elif 'friend request' in cell:
return 'friend request'
elif 'profile picture' in cell:
return 'profile picture'
elif 'an event' in cell:
return 'an event'
elif 'a list' in cell:
return 'a list'
elif 'an entry' in cell:
return 'an entry'
elif 'a note' in cell:
return 'a note'
elif 'Quotations' in cell:
return 'quotations'
elif 'phone number' in cell:
return 'phone number'
elif 'Website' in cell:
return 'website'
elif 'memory' in cell:
return 'memory'
elif 'poll' in cell:
return 'poll'
elif 'hometown' in cell:
return 'hometown'
elif 'Page' in cell:
return 'page'
else:
return None
people2 = pd.DataFrame(people2)
# people2[~(people2[0].map(parse_comments)==people2[0].map(parse_comments))]
people2['media'] = people2[0].map(parse_comments)
#top media that you interact with
people2.media.value_counts()[:10]
people2.rename(columns={0:'event_text'},inplace=True)
def parse_action(cell):
if 'liked' in cell:
return 'liked'
elif 'likes' in cell:
return 'likes'
elif 'reacted to' in cell:
return 'reacted to'
elif 'is going to' in cell:
return 'is going to'
elif 'was mentioned in a' in cell:
return 'was mentioned in a'
elif 'commented on' in cell:
return 'commented on'
elif 'posted in' in cell:
return 'posted in'
elif 'became friends with' in cell:
return 'became friends with'
elif 'replied to' in cell:
return 'replied to'
elif 'added' in cell:
return 'added'
elif 'shared a post to your' in cell:
return 'shared a post to your'
elif 'was tagged in' in cell:
return 'was tagged in'
elif 'shared' in cell:
return 'shared'
elif 'wrote on your' in cell:
return 'wrote on your'
elif 'wrote on' in cell:
return 'wrote on'
elif 'updated' in cell:
return 'updated'
elif 'sent' in cell:
return 'sent'
elif 'changed' in cell:
return 'changed'
elif 'posted a video to' in cell:
return 'posted a video to'
elif 'posted something via' in cell:
return 'posted something via'
elif 'voted for' in cell:
return 'voted for'
elif 'was feeling' in cell:
return 'was feeling'
elif 'reviewed' in cell:
return 'reviewed'
elif 'created' in cell:
return 'created'
elif 'was tagged at' in cell:
return 'was tagged at'
elif 'voted on' in cell:
return 'voted on'
elif 'was with' in cell:
return 'was with'
elif 'is now using Facebook in' in cell:
return 'is now using Facebook in'
elif 'is interested in' in cell:
return 'is interested in'
elif 'was' in cell:
return 'was'
# people2[~(people2.event_text.map(parse_action)==people2.event_text.map(parse_action))]
people2['event'] = people2.event_text.map(parse_action)
#top interactions on Facebook
people2.event.value_counts()[:10]
people2['date'] = people2.event_text.map(lambda x: x.split('.')[-1])
for x in people2.index:
if people2.loc[x]['event_text'] != people2.loc[x]['event_text']:
people2.loc[x,'name'] = None
else:
test = people2.loc[x]['event_text']
if people2.loc[x,'event'] == 'became friends with':
person = test[test.find(people2.loc[x]['event']) + len(people2.loc[x]['event']) + 1:test.find('.')]
people2.loc[x,'name'] = person
elif ' own ' in people2.loc[x,'event_text'] or ' his ' in people2.loc[x,'event_text'] or ' her ' in people2.loc[x,'event_text']:
people2.loc[x,'name'] = None
elif people2.loc[x,'event'] == 'added':
people2.loc[x,'name'] = None
elif people2.loc[x,'media'] == None:
people2.loc[x,'name'] = None
else:
person = test[test.find(people2.loc[x]['event']) + len(people2.loc[x]['event']) + 1:test.find(people2.loc[x]['media']) - 3]
people2.loc[x,'name'] = person
#people you interact with the most
people2[people2.name != ''].name.value_counts()[:20]
#things you interact with the most
people2.media.value_counts()[:10]
posts2 = []
for x in posts:
posts2.append(x.text)
text = ' '.join(posts2)
text2 = text.replace('\n\n',' ').replace('\n',' ').replace(u"\'","").lower()
test = pd.Series(text2.split(' '))
urls = test[test.str.contains('http')|test.str.contains('www')].values
test = test[~(test.str.contains('http')|test.str.contains('www')|test.str.contains('.com'))].copy()
text2 = test.str.cat(sep=' ')
import string
for x in list(string.punctuation):
text2 = text2.replace(x,' ')
words_list = text2.split(' ')
words_list = pd.Series(words_list)
words_list = words_list[words_list != ''].copy()
stop = stopwords.words('english')
final_words_list = words_list[~words_list.isin(stop)]
#words in Facebook interactions you have had
final_words_list.value_counts()
#look at likes and comments over time for a single post (top liked and commented posts--to get a sense for timeline of liking and posting)
#when do I like, comment, or post things? Any patterns?
#look at URLs I have posted about (top URLs...?)
#look at hashtags I have used (how am I treating #aslkdflksd--I need to pull them out)
| 0.254139 | 0.234801 |
# Using `pybind11`
The package `pybind11` is provides an elegant way to wrap C++ code for Python, including automatic conversions for `numpy` arrays and the C++ `Eigen` linear algebra library. Used with the `cppimport` package, this provides a very nice work flow for integrating C++ and Python:
- Edit C++ code
- Run Python code
```bash
! pip install pybind11
! pip install cppimport
```
Clone the Eigen library if necessary - no installation is required as Eigen is a header only library.
```bash
! git clone https://github.com/RLovelett/eigen.git
```
## Resources
- [`pybind11`](http://pybind11.readthedocs.io/en/latest/)
- [`cppimport`](https://github.com/tbenthompson/cppimport)
- [`Eigen`](http://eigen.tuxfamily.org)
## A first example of using `pybind11`
Create a new subdirectory - e.g. `example1` and create the following 5 files in it:
- `funcs.hpp`
- `funcs.cpp`
- `wrap.cpp`
- `setup.py`
- `test_funcs.py`
First write the C++ header and implementation files
```
%mkdir example1
%cd example1
%%file funcs.hpp
int add(int i, int j);
%%file funcs.cpp
int add(int i, int j) {
return i + j;
};
```
Next write the C++ wrapper code using `pybind11` in `wrap.cpp`. The arguments `"i"_a=1, "j"_a=2` in the exported function definition tells `pybind11` to generate variables named `i` with default value 1 and `j` with default value 2 for the `add` function.
```
%%file wrap1.cpp
#include <pybind11/pybind11.h>
#include "funcs.hpp"
namespace py = pybind11;
using namespace pybind11::literals;
PYBIND11_MODULE(wrap1, m) {
m.doc() = "pybind11 example plugin";
m.def("add", &add, "A function which adds two numbers",
"i"_a=1, "j"_a=2);
}
```
Finally, write the `setup.py` file to compile the extension module. This is mostly boilerplate.
```
%%file setup.py
import os, sys
from distutils.core import setup, Extension
from distutils import sysconfig
cpp_args = ['-std=c++11']
ext_modules = [
Extension(
'wrap1',
['funcs.cpp', 'wrap1.cpp'],
include_dirs=['pybind11/include'],
language='c++',
extra_compile_args = cpp_args,
),
]
setup(
name='wrap1',
version='0.0.1',
author='Cliburn Chan',
author_email='cliburn.chan@duke.edu',
description='Example',
ext_modules=ext_modules,
)
```
Now build the extension module in the subdirectory with these files
```
%%bash
python setup.py build_ext -i
```
And if you are successful, you should now see a new ```funcs.so``` extension module. We can write a `test_funcs.py` file test the extension module:
```
%%file test_funcs.py
import wrap1
def test_add():
print(wrap1.add(3, 4))
assert(wrap1.add(3, 4) == 7)
if __name__ == '__main__':
test_add()
```
And finally, running the test should not generate any error messages:
```
%%bash
python test_funcs.py
%cd ..
```
## Using `cppimport`
In the development stage, it can be distracting to have to repeatedly rebuild the extension module by running
```bash
python setup.py clean
python setup.py build_ext -i
```
every single time you modify the C++ code. The `cppimport` package does this for you.
Create a new sub-directory `exaample2` and copy the files `func.hpp`, `funcs.cpp` and `wrap.cpp` from `example1` over.
For the previous example, we just need to add some annotation (between `<% and %>` delimiters) to the top of the `wrap.cpp` file
```
%mkdir example2
%cp example1/funcs.* example2/
%cd example2
%%file wrap2.cpp
<%
cfg['compiler_args'] = ['-std=c++11']
cfg['sources'] = ['funcs.cpp']
setup_pybind11(cfg)
%>
#include "funcs.hpp"
#include <pybind11/pybind11.h>
namespace py = pybind11;
PYBIND11_MODULE(wrap2, m) {
m.doc() = "pybind11 example plugin";
m.def("add", &add, "A function which adds two numbers");
}
%%file test_funcs.py
import cppimport
funcs = cppimport.imp("wrap2")
def test_add():
assert(funcs.add(3, 4) == 7)
if __name__ == '__main__':
print(funcs.add(3,4))
test_add()
%%bash
python test_funcs.py
```
### Use of `cppimport`
Note that `cppimport.imp` is only called once. Once it is called, the shared library is created and can be sued
```
! ls *so
```
That is, you can import wrap2 and call from notebook
```
import wrap2
wrap2.add(3, 4)
```
without any need to manually build the extension module. Any updates will be detected by `cppimport` and it will automatically trigger a re-build.
```
%cd ..
```
## Vectorizing functions for use with `numpy` arrays
Example showing how to vectorize a `square` function. Note that from here on, we don't bother to use separate header and implementation files for these code snippets, and just write them together with the wrapping code in a `code.cpp` file. This means that with `cppimport`, there are only two files that we actually code for, a C++ `code.cpp` file and a python test file.
```
%mkdir example3
%cd example3
%%file wrap3.cpp
<%
cfg['compiler_args'] = ['-std=c++11']
setup_pybind11(cfg)
%>
#include <pybind11/pybind11.h>
#include <pybind11/numpy.h>
namespace py = pybind11;
double square(double x) {
return x * x;
}
PYBIND11_MODULE(wrap3, m) {
m.doc() = "pybind11 example plugin";
m.def("square", py::vectorize(square), "A vectroized square function.");
}
import cppimport
wrap3 = cppimport.imp("wrap3")
wrap3.square([1,2,3])
```
Once the shared libary is built, you can use it as a regular Python module.
```
! ls
import wrap3
wrap3.square([2,4,6])
%cd ..
```
## Using `numpy` arrays as function arguments and return values
Example showing how to pass `numpy` arrays in and out of functions. These `numpy` array arguments can either be generic `py:array` or typed `py:array_t<double>`. The properties of the `numpy` array can be obtained by calling its `request` method. This returns a `struct` of the following form:
```c++
struct buffer_info {
void *ptr;
size_t itemsize;
std::string format;
int ndim;
std::vector<size_t> shape;
std::vector<size_t> strides;
};
```
Here is C++ code for two functions - the function `twice` shows how to change a passed in `numpy` array in-place using pointers; the function `sum` shows how to sum the elements of a `numpy` array. By taking advantage of the information in `buffer_info`, the code will work for arbitrary `n-d` arrays.
```
%mkdir example4
%cd example4
%%file wrap4.cpp
<%
cfg['compiler_args'] = ['-std=c++11']
setup_pybind11(cfg)
%>
#include <pybind11/pybind11.h>
#include <pybind11/numpy.h>
namespace py = pybind11;
// Passing in an array of doubles
void twice(py::array_t<double> xs) {
py::buffer_info info = xs.request();
auto ptr = static_cast<double *>(info.ptr);
int n = 1;
for (auto r: info.shape) {
n *= r;
}
for (int i = 0; i <n; i++) {
*ptr++ *= 2;
}
}
// Passing in a generic array
double sum(py::array xs) {
py::buffer_info info = xs.request();
auto ptr = static_cast<double *>(info.ptr);
int n = 1;
for (auto r: info.shape) {
n *= r;
}
double s = 0.0;
for (int i = 0; i <n; i++) {
s += *ptr++;
}
return s;
}
PYBIND11_MODULE(wrap4, m) {
m.doc() = "auto-compiled c++ extension";
m.def("sum", &sum);
m.def("twice", &twice);
}
%%file test_code.py
import cppimport
import numpy as np
code = cppimport.imp("wrap4")
if __name__ == '__main__':
xs = np.arange(12).reshape(3,4).astype('float')
print(xs)
print("np :", xs.sum())
print("cpp:", code.sum(xs))
print()
code.twice(xs)
print(xs)
%%bash
python test_code.py
%cd ..
```
## More on working with `numpy` arrays
This example shows how to use array access for `numpy` arrays within the C++ function. It is taken from the `pybind11` documentation, but fixes a small bug in the official version. As noted in the documentation, the function would be more easily coded using `py::vectorize`.
```
%mkdir example5
%cd example5
%%file wrap5.cpp
<%
cfg['compiler_args'] = ['-std=c++11']
setup_pybind11(cfg)
%>
#include <pybind11/pybind11.h>
#include <pybind11/numpy.h>
namespace py = pybind11;
py::array_t<double> add_arrays(py::array_t<double> input1, py::array_t<double> input2) {
auto buf1 = input1.request(), buf2 = input2.request();
if (buf1.ndim != 1 || buf2.ndim != 1)
throw std::runtime_error("Number of dimensions must be one");
if (buf1.shape[0] != buf2.shape[0])
throw std::runtime_error("Input shapes must match");
auto result = py::array(py::buffer_info(
nullptr, /* Pointer to data (nullptr -> ask NumPy to allocate!) */
sizeof(double), /* Size of one item */
py::format_descriptor<double>::value, /* Buffer format */
buf1.ndim, /* How many dimensions? */
{ buf1.shape[0] }, /* Number of elements for each dimension */
{ sizeof(double) } /* Strides for each dimension */
));
auto buf3 = result.request();
double *ptr1 = (double *) buf1.ptr,
*ptr2 = (double *) buf2.ptr,
*ptr3 = (double *) buf3.ptr;
for (size_t idx = 0; idx < buf1.shape[0]; idx++)
ptr3[idx] = ptr1[idx] + ptr2[idx];
return result;
}
PYBIND11_MODULE(wrap5, m) {
m.def("add_arrays", &add_arrays, "Add two NumPy arrays");
}
import cppimport
import numpy as np
code = cppimport.imp("wrap5")
xs = np.arange(12)
print(xs)
print(code.add_arrays(xs, xs))
%cd ..
```
## Using the C++ `eigen` library to calculate matrix inverse and determinant
Example showing how `Eigen` vectors and matrices can be passed in and out of C++ functions. Note that `Eigen` arrays are automatically converted to/from `numpy` arrays simply by including the `pybind/eigen.h` header. Because of this, it is probably simplest in most cases to work with `Eigen` vectors and matrices rather than `py::buffer` or `py::array` where `py::vectorize` is insufficient.
```
%mkdir example6
%cd example6
%%file wrap6.cpp
<%
cfg['compiler_args'] = ['-std=c++11']
cfg['include_dirs'] = ['../eigen3']
setup_pybind11(cfg)
%>
#include <pybind11/pybind11.h>
#include <pybind11/eigen.h>
#include <Eigen/LU>
namespace py = pybind11;
// convenient matrix indexing comes for free
double get(Eigen::MatrixXd xs, int i, int j) {
return xs(i, j);
}
// takes numpy array as input and returns double
double det(Eigen::MatrixXd xs) {
return xs.determinant();
}
// takes numpy array as input and returns another numpy array
Eigen::MatrixXd inv(Eigen::MatrixXd xs) {
return xs.inverse();
}
PYBIND11_MODULE(wrap6, m) {
m.doc() = "auto-compiled c++ extension";
m.def("inv", &inv);
m.def("det", &det);
}
import cppimport
import numpy as np
code = cppimport.imp("wrap6")
A = np.array([[1,2,1],
[2,1,0],
[-1,1,2]])
print(A)
print(code.det(A))
print(code.inv(A))
%cd ..
```
## Using `pybind11` with `openmp`
```
%mkdir example7
%cd example7
```
Here is an example of using OpenMP to integrate the value of $\pi$ written using `pybind11`.
```
%%file wrap7.cpp
/*
<%
cfg['compiler_args'] = ['-std=c++11', '-fopenmp']
cfg['linker_args'] = ['-lgomp']
setup_pybind11(cfg)
%>
*/
#include <cmath>
#include <omp.h>
#include <pybind11/pybind11.h>
#include <pybind11/numpy.h>
namespace py = pybind11;
// Passing in an array of doubles
void twice(py::array_t<double> xs) {
py::gil_scoped_acquire acquire;
py::buffer_info info = xs.request();
auto ptr = static_cast<double *>(info.ptr);
int n = 1;
for (auto r: info.shape) {
n *= r;
}
#pragma omp parallel for
for (int i = 0; i <n; i++) {
*ptr++ *= 2;
}
}
PYBIND11_MODULE(wrap7, m) {
m.doc() = "auto-compiled c++ extension";
m.def("twice", [](py::array_t<double> xs) {
/* Release GIL before calling into C++ code */
py::gil_scoped_release release;
return twice(xs);
});
}
import cppimport
import numpy as np
code = cppimport.imp("wrap7")
xs = np.arange(10).astype('double')
code.twice(xs)
xs
%cd ..
```
|
github_jupyter
|
! pip install pybind11
! pip install cppimport
! git clone https://github.com/RLovelett/eigen.git
%mkdir example1
%cd example1
%%file funcs.hpp
int add(int i, int j);
%%file funcs.cpp
int add(int i, int j) {
return i + j;
};
%%file wrap1.cpp
#include <pybind11/pybind11.h>
#include "funcs.hpp"
namespace py = pybind11;
using namespace pybind11::literals;
PYBIND11_MODULE(wrap1, m) {
m.doc() = "pybind11 example plugin";
m.def("add", &add, "A function which adds two numbers",
"i"_a=1, "j"_a=2);
}
%%file setup.py
import os, sys
from distutils.core import setup, Extension
from distutils import sysconfig
cpp_args = ['-std=c++11']
ext_modules = [
Extension(
'wrap1',
['funcs.cpp', 'wrap1.cpp'],
include_dirs=['pybind11/include'],
language='c++',
extra_compile_args = cpp_args,
),
]
setup(
name='wrap1',
version='0.0.1',
author='Cliburn Chan',
author_email='cliburn.chan@duke.edu',
description='Example',
ext_modules=ext_modules,
)
%%bash
python setup.py build_ext -i
%%file test_funcs.py
import wrap1
def test_add():
print(wrap1.add(3, 4))
assert(wrap1.add(3, 4) == 7)
if __name__ == '__main__':
test_add()
%%bash
python test_funcs.py
%cd ..
python setup.py clean
python setup.py build_ext -i
%mkdir example2
%cp example1/funcs.* example2/
%cd example2
%%file wrap2.cpp
<%
cfg['compiler_args'] = ['-std=c++11']
cfg['sources'] = ['funcs.cpp']
setup_pybind11(cfg)
%>
#include "funcs.hpp"
#include <pybind11/pybind11.h>
namespace py = pybind11;
PYBIND11_MODULE(wrap2, m) {
m.doc() = "pybind11 example plugin";
m.def("add", &add, "A function which adds two numbers");
}
%%file test_funcs.py
import cppimport
funcs = cppimport.imp("wrap2")
def test_add():
assert(funcs.add(3, 4) == 7)
if __name__ == '__main__':
print(funcs.add(3,4))
test_add()
%%bash
python test_funcs.py
! ls *so
import wrap2
wrap2.add(3, 4)
%cd ..
%mkdir example3
%cd example3
%%file wrap3.cpp
<%
cfg['compiler_args'] = ['-std=c++11']
setup_pybind11(cfg)
%>
#include <pybind11/pybind11.h>
#include <pybind11/numpy.h>
namespace py = pybind11;
double square(double x) {
return x * x;
}
PYBIND11_MODULE(wrap3, m) {
m.doc() = "pybind11 example plugin";
m.def("square", py::vectorize(square), "A vectroized square function.");
}
import cppimport
wrap3 = cppimport.imp("wrap3")
wrap3.square([1,2,3])
! ls
import wrap3
wrap3.square([2,4,6])
%cd ..
Here is C++ code for two functions - the function `twice` shows how to change a passed in `numpy` array in-place using pointers; the function `sum` shows how to sum the elements of a `numpy` array. By taking advantage of the information in `buffer_info`, the code will work for arbitrary `n-d` arrays.
## More on working with `numpy` arrays
This example shows how to use array access for `numpy` arrays within the C++ function. It is taken from the `pybind11` documentation, but fixes a small bug in the official version. As noted in the documentation, the function would be more easily coded using `py::vectorize`.
## Using the C++ `eigen` library to calculate matrix inverse and determinant
Example showing how `Eigen` vectors and matrices can be passed in and out of C++ functions. Note that `Eigen` arrays are automatically converted to/from `numpy` arrays simply by including the `pybind/eigen.h` header. Because of this, it is probably simplest in most cases to work with `Eigen` vectors and matrices rather than `py::buffer` or `py::array` where `py::vectorize` is insufficient.
## Using `pybind11` with `openmp`
Here is an example of using OpenMP to integrate the value of $\pi$ written using `pybind11`.
| 0.493164 | 0.921922 |
# Continuous training with TFX and Vertex
## Learning Objectives
1. Containerize your TFX code into a pipeline package using Cloud Build.
1. Use the TFX CLI to compile a TFX pipeline.
1. Deploy a TFX pipeline version to run on Vertex Pipelines using the Vertex Python SDK.
### Setup
```
from google.cloud import aiplatform as vertex_ai
```
#### Validate lab package version installation
```
!python -c "import tensorflow as tf; print(f'TF version: {tf.__version__}')"
!python -c "import tfx; print(f'TFX version: {tfx.__version__}')"
!python -c "import kfp; print(f'KFP version: {kfp.__version__}')"
print(f"aiplatform: {vertex_ai.__version__}")
```
**Note**: this lab was built and tested with the following package versions:
`TF version: 2.6.2`
`TFX version: 1.4.0`
`KFP version: 1.8.1`
`aiplatform: 1.7.1`
## Review: example TFX pipeline design pattern for Vertex
The pipeline source code can be found in the `pipeline_vertex` folder.
```
%cd pipeline_vertex
!ls -la
```
The `config.py` module configures the default values for the environment specific settings and the default values for the pipeline runtime parameters.
The default values can be overwritten at compile time by providing the updated values in a set of environment variables. You will set custom environment variables later on this lab.
The `pipeline.py` module contains the TFX DSL defining the workflow implemented by the pipeline.
The `preprocessing.py` module implements the data preprocessing logic the `Transform` component.
The `model.py` module implements the TensorFlow model code and training logic for the `Trainer` component.
The `runner.py` module configures and executes `KubeflowV2DagRunner`. At compile time, the `KubeflowDagRunner.run()` method converts the TFX DSL into the pipeline package into a JSON format for execution on Vertex.
The `features.py` module contains feature definitions common across `preprocessing.py` and `model.py`.
## Exercise: build your pipeline with the TFX CLI
You will use TFX CLI to compile and deploy the pipeline. As explained in the previous section, the environment specific settings can be provided through a set of environment variables and embedded into the pipeline package at compile time.
### Configure your environment resource settings
Update the below constants with the settings reflecting your lab environment.
- `REGION` - the compute region for AI Platform Training, Vizier, and Prediction.
- `ARTIFACT_STORE` - An existing GCS bucket. You can use any bucket, but we will use here the bucket with the same name as the project.
```
# TODO: Set your environment resource settings here for GCP_REGION, ARTIFACT_STORE_URI, ENDPOINT, and CUSTOM_SERVICE_ACCOUNT.
REGION = "us-central1"
PROJECT_ID = !(gcloud config get-value core/project)
PROJECT_ID = PROJECT_ID[0]
ARTIFACT_STORE = f"gs://{PROJECT_ID}"
# Set your resource settings as environment variables. These override the default values in pipeline/config.py.
%env REGION={REGION}
%env ARTIFACT_STORE={ARTIFACT_STORE}
%env PROJECT_ID={PROJECT_ID}
!gsutil ls | grep ^{ARTIFACT_STORE}/$ || gsutil mb -l {REGION} {ARTIFACT_STORE}
```
### Set the compile time settings to first create a pipeline version without hyperparameter tuning
Default pipeline runtime environment values are configured in the pipeline folder `config.py`. You will set their values directly below:
* `PIPELINE_NAME` - the pipeline's globally unique name.
* `DATA_ROOT_URI` - the URI for the raw lab dataset `gs://{PROJECT_ID}/data/tfxcovertype`.
* `TFX_IMAGE_URI` - the image name of your pipeline container that will be used to execute each of your tfx components
```
PIPELINE_NAME = "tfxcovertype"
DATA_ROOT_URI = f"gs://{PROJECT_ID}/data/tfxcovertype"
TFX_IMAGE_URI = f"gcr.io/{PROJECT_ID}/{PIPELINE_NAME}"
PIPELINE_JSON = f"{PIPELINE_NAME}.json"
TRAIN_STEPS = 10
EVAL_STEPS = 5
%env PIPELINE_NAME={PIPELINE_NAME}
%env DATA_ROOT_URI={DATA_ROOT_URI}
%env TFX_IMAGE_URI={TFX_IMAGE_URI}
%env PIPELINE_JSON={PIPELINE_JSON}
%env TRAIN_STEPS={TRAIN_STEPS}
%env EVAL_STEPS={EVAL_STEPS}
```
Let us populate the data bucket at `DATA_ROOT_URI`:
```
!gsutil cp ../../../data/* $DATA_ROOT_URI/dataset.csv
!gsutil ls $DATA_ROOT_URI/*
```
Let us build and push the TFX container image described in the `Dockerfile`:
```
!gcloud builds submit --timeout 15m --tag $TFX_IMAGE_URI .
```
### Compile your pipeline code
The following command will execute the `KubeflowV2DagRunner` that compiles the pipeline described in `pipeline.py` into a JSON representation consumable by Vertex:
```
!tfx pipeline compile --engine vertex --pipeline_path runner.py
```
Note: you should see a `{PIPELINE_NAME}.json` file appear in your current pipeline directory.
## Exercise: deploy your pipeline on Vertex using the Vertex SDK
Once you have the `{PIPELINE_NAME}.json` available, you can run the tfx pipeline on Vertex by launching a pipeline job using the `aiplatform` handle:
```
from google.cloud import aiplatform as vertex_ai
vertex_ai.init(project=PROJECT_ID,location=REGION)
pipeline = vertex_ai.PipelineJob(
display_name="mypiplineTFX",
template_path=PIPELINE_JSON,
enable_caching=False,
)
pipeline.run(service_account="qwiklabs-gcp-01-9a9d18213c32@qwiklabs-gcp-01-9a9d18213c32.iam.gserviceaccount.com")
```
## Next Steps
In this lab, you learned how to build and deploy a TFX pipeline with the TFX CLI and then update, build and deploy a new pipeline with automatic hyperparameter tuning. You practiced triggered continuous pipeline runs using the TFX CLI as well as the Kubeflow Pipelines UI.
In the next lab, you will construct a Cloud Build CI/CD workflow that further automates the building and deployment of the pipeline.
## License
Copyright 2021 Google Inc. All Rights Reserved.
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
|
github_jupyter
|
from google.cloud import aiplatform as vertex_ai
!python -c "import tensorflow as tf; print(f'TF version: {tf.__version__}')"
!python -c "import tfx; print(f'TFX version: {tfx.__version__}')"
!python -c "import kfp; print(f'KFP version: {kfp.__version__}')"
print(f"aiplatform: {vertex_ai.__version__}")
%cd pipeline_vertex
!ls -la
# TODO: Set your environment resource settings here for GCP_REGION, ARTIFACT_STORE_URI, ENDPOINT, and CUSTOM_SERVICE_ACCOUNT.
REGION = "us-central1"
PROJECT_ID = !(gcloud config get-value core/project)
PROJECT_ID = PROJECT_ID[0]
ARTIFACT_STORE = f"gs://{PROJECT_ID}"
# Set your resource settings as environment variables. These override the default values in pipeline/config.py.
%env REGION={REGION}
%env ARTIFACT_STORE={ARTIFACT_STORE}
%env PROJECT_ID={PROJECT_ID}
!gsutil ls | grep ^{ARTIFACT_STORE}/$ || gsutil mb -l {REGION} {ARTIFACT_STORE}
PIPELINE_NAME = "tfxcovertype"
DATA_ROOT_URI = f"gs://{PROJECT_ID}/data/tfxcovertype"
TFX_IMAGE_URI = f"gcr.io/{PROJECT_ID}/{PIPELINE_NAME}"
PIPELINE_JSON = f"{PIPELINE_NAME}.json"
TRAIN_STEPS = 10
EVAL_STEPS = 5
%env PIPELINE_NAME={PIPELINE_NAME}
%env DATA_ROOT_URI={DATA_ROOT_URI}
%env TFX_IMAGE_URI={TFX_IMAGE_URI}
%env PIPELINE_JSON={PIPELINE_JSON}
%env TRAIN_STEPS={TRAIN_STEPS}
%env EVAL_STEPS={EVAL_STEPS}
!gsutil cp ../../../data/* $DATA_ROOT_URI/dataset.csv
!gsutil ls $DATA_ROOT_URI/*
!gcloud builds submit --timeout 15m --tag $TFX_IMAGE_URI .
!tfx pipeline compile --engine vertex --pipeline_path runner.py
from google.cloud import aiplatform as vertex_ai
vertex_ai.init(project=PROJECT_ID,location=REGION)
pipeline = vertex_ai.PipelineJob(
display_name="mypiplineTFX",
template_path=PIPELINE_JSON,
enable_caching=False,
)
pipeline.run(service_account="qwiklabs-gcp-01-9a9d18213c32@qwiklabs-gcp-01-9a9d18213c32.iam.gserviceaccount.com")
| 0.250638 | 0.945147 |
# Quitar Overfit a Decision Tree
La implementación por defecto de DecisionTreeClassifier de Scikit-learn hace overfit, pues no pone ningún límite al crecimiento del árbol. Pero no está claro hasta qué punto tenemos que dejarlo crecer, y hay muchos hiper-parámetros para modificar esto. En este notebook voy a trastear con los parámetros para ver como se comportan
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from time import time
import math
# Import datasets, classifiers and performance metrics
from sklearn import datasets
from sklearn.tree import DecisionTreeClassifier
```
## min_impurity_decrease
```
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
#decreases = np.arange(0,100)
#decreases = np.arange(0,0.1, 0.01)
decreases = np.linspace(0,0.1)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dec in decreases:
clf.set_params(min_impurity_decrease = dec)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(decreases, test_scores, label = "Test score")
accuracy.plot(decreases, train_scores, label = "Train score")
#accuracy.xaxis.set_ticks(np.arange(0,100, 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("min impurity decrease")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
```
min_impurity_decrease
A node will be split if this split induces a decrease of the impurity greater than or equal to this value.
The weighted impurity decrease equation is the following:
```
N_t / N * (impurity - N_t_R / N_t * right_impurity
- N_t_L / N_t * left_impurity)
```
## max_depth
```
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
depth = np.arange(1,100)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dep in depth:
clf.set_params(max_depth = dep)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(depth, test_scores, label = "Test score")
accuracy.plot(depth, train_scores, label = "Train score")
#accuracy.xaxis.set_ticks(np.arange(0,depth[-1], depth[-1] / 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("Max depth")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
```
max_depth
The maximum depth of the tree. If None, then nodes are expanded until all leaves are pure or until all leaves contain less than min_samples_split samples
## min_samples_split
```
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
sample_sizes = np.arange(2,150)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dep in sample_sizes:
clf.set_params(min_samples_split = dep)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(sample_sizes, test_scores, label = "Test score")
accuracy.plot(sample_sizes, train_scores, label = "Train score")
#accuracy.xaxis.set_ticks(np.arange(0,depth[-1], depth[-1] / 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("min_samples_split")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
```
min_samples_split
The minimum number of samples required to split an internal node
## min_samples_leaf
```
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
sample_sizes = np.arange(1,150)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dep in sample_sizes:
clf.set_params(min_samples_leaf = dep)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(sample_sizes, test_scores, label = "Test Score")
accuracy.plot(sample_sizes, train_scores, label = "Train Score")
#accuracy.xaxis.set_ticks(np.arange(0,depth[-1], depth[-1] / 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("min_samples_leaf")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
```
min_samples_leaf
The minimum number of samples required to be at a leaf node. A split point at any depth will only be considered if it leaves at least min_samples_leaf training samples in each of the left and right branches. This may have the effect of smoothing the model, especially in regression.
## min_weight_fraction_leaf
```
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
#sample_sizes = np.arange(0.0,0.5, 0.02)
sample_sizes = np.linspace(0,0.5)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dep in sample_sizes:
clf.set_params(min_weight_fraction_leaf = dep)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(sample_sizes, test_scores, label = "Test Score")
accuracy.plot(sample_sizes, train_scores, label = "Train Score")
#accuracy.xaxis.set_ticks(np.arange(0,depth[-1], depth[-1] / 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("min_weight_fraction_leaf")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
```
min_weight_fraction_leaf
The minimum weighted fraction of the sum total of weights (of all the input samples) required to be at a leaf node. Samples have equal weight when sample_weight is not provided.
## max_leaf_nodes
```
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
sample_sizes = np.arange(2, 200)
#sample_sizes = np.linspace(0,0.5)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dep in sample_sizes:
clf.set_params(max_leaf_nodes = dep)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(sample_sizes, test_scores, label = "Test Score")
accuracy.plot(sample_sizes, train_scores, label = "Train Score")
#accuracy.xaxis.set_ticks(np.arange(0,depth[-1], depth[-1] / 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("max_leaf_nodes")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
```
max_leaf_nodes
Grow a tree with max_leaf_nodes in best-first fashion. Best nodes are defined as relative reduction in impurity. If None then unlimited number of leaf nodes.
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from time import time
import math
# Import datasets, classifiers and performance metrics
from sklearn import datasets
from sklearn.tree import DecisionTreeClassifier
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
#decreases = np.arange(0,100)
#decreases = np.arange(0,0.1, 0.01)
decreases = np.linspace(0,0.1)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dec in decreases:
clf.set_params(min_impurity_decrease = dec)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(decreases, test_scores, label = "Test score")
accuracy.plot(decreases, train_scores, label = "Train score")
#accuracy.xaxis.set_ticks(np.arange(0,100, 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("min impurity decrease")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
N_t / N * (impurity - N_t_R / N_t * right_impurity
- N_t_L / N_t * left_impurity)
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
depth = np.arange(1,100)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dep in depth:
clf.set_params(max_depth = dep)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(depth, test_scores, label = "Test score")
accuracy.plot(depth, train_scores, label = "Train score")
#accuracy.xaxis.set_ticks(np.arange(0,depth[-1], depth[-1] / 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("Max depth")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
sample_sizes = np.arange(2,150)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dep in sample_sizes:
clf.set_params(min_samples_split = dep)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(sample_sizes, test_scores, label = "Test score")
accuracy.plot(sample_sizes, train_scores, label = "Train score")
#accuracy.xaxis.set_ticks(np.arange(0,depth[-1], depth[-1] / 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("min_samples_split")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
sample_sizes = np.arange(1,150)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dep in sample_sizes:
clf.set_params(min_samples_leaf = dep)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(sample_sizes, test_scores, label = "Test Score")
accuracy.plot(sample_sizes, train_scores, label = "Train Score")
#accuracy.xaxis.set_ticks(np.arange(0,depth[-1], depth[-1] / 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("min_samples_leaf")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
#sample_sizes = np.arange(0.0,0.5, 0.02)
sample_sizes = np.linspace(0,0.5)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dep in sample_sizes:
clf.set_params(min_weight_fraction_leaf = dep)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(sample_sizes, test_scores, label = "Test Score")
accuracy.plot(sample_sizes, train_scores, label = "Train Score")
#accuracy.xaxis.set_ticks(np.arange(0,depth[-1], depth[-1] / 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("min_weight_fraction_leaf")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
digits = datasets.load_digits()
data = digits.data
target = digits.target
N = data.shape[0]
prop_train = 2 / 3
N_train = math.ceil(N * prop_train)
N_test = N - N_train
data = digits.data
# Algunas columnas tienen todo 0. Estas features no aportan ninguna información,
# y además me impiden estandarizar, pues la std es 0
# La vamos a quitar. Este array es de booleanos
valid_cols = np.apply_along_axis(lambda a: np.count_nonzero(a) > 0, axis = 0, arr = data)
data.shape
data = data[:, valid_cols]
data.shape
mean = data.mean(axis = 0)
std = data.std(axis = 0)
data = (data - mean)/std
data_train = data[:N_train]
data_test = data[N_train:]
target_train = target[:N_train]
target_test = target[N_train:]
sample_sizes = np.arange(2, 200)
#sample_sizes = np.linspace(0,0.5)
test_scores = []
train_scores = []
clf = DecisionTreeClassifier()
for dep in sample_sizes:
clf.set_params(max_leaf_nodes = dep)
clf.fit(data_train, target_train)
test_score = clf.score(data_test, target_test)
train_score = clf.score(data_train, target_train)
test_scores.append(test_score)
train_scores.append(train_score)
accuracy = plt.subplot(111)
accuracy.plot(sample_sizes, test_scores, label = "Test Score")
accuracy.plot(sample_sizes, train_scores, label = "Train Score")
#accuracy.xaxis.set_ticks(np.arange(0,depth[-1], depth[-1] / 10))
accuracy.locator_params(nbins = 15, axis = "x")
accuracy.grid(True)
plt.xlabel("max_leaf_nodes")
plt.ylabel("Accuracy")
accuracy.legend(loc='best')
| 0.36523 | 0.944485 |
###Beginner Tutorial: Neural Networks in Theano
####What is Theano and why should I use it?
Theano is part framework and part library for evaluating and optimizing mathematical expressions. It's popular in the machine learning world because it allows you to build up optimized symbolic computational graphs and the gradients can be automatically computed. Moreover, Theano also supports running code on the GPU. Automatic gradients + GPU sounds pretty nice. I won't be showing you how to run on the GPU because I'm using a Macbook Air and as far as I know, Theano doesn't support or barely supports OpenCL at this time. But you can check out their <a href="http://deeplearning.net/software/theano/tutorial/using_gpu.html">documentation</a> if you have an nVidia GPU ready to go.
####Summary
As the title suggests, I'm going to show how to build a simple neural network (yep, you guessed it, using our favorite XOR problem..) using Theano. The reason I wrote this post is because I found the existing Theano tutorials to be not simple enough. I'm all about reducing things to fundamentals. Given that, I will not be using all the bells-and-whistles that Theano has to offer and I'm going to be writing code that maximizes for readability. Nonetheless, using what I show here, you should be able to scale up to more complex algorithms.
####Assumptions
I assume you know how to write a simple neural network in Python (including training it with gradient descent/backpropagation). I also assume you've at least browsed through the Theano <a href="http://deeplearning.net/software/theano/index.html">documentation</a> and have a feel for what it's about (I didn't do it justice in my explanation of "why Theano" above).
###Let's get started
First, let's import all the goodies we'll need.
```
import theano
import theano.tensor as T
import theano.tensor.nnet as nnet
import numpy as np
```
Before we actually build the neural network, let's just get familiarized with how Theano works. Let's do something really simple, we'll simply ask Theano to give us the derivative of a simple mathematical expression like
$$ f(x) = e^{sin{(x^2)}} $$
As you can see, this is an equation of a single variable $x$. So let's use Theano to symbolically define our variable $x$. What do I mean by symbolically? Well, we're going to be building a Theano expression using variables and numbers similar to how we'd write this equation down on paper. We're not actually computing anything yet. Since Theano is a Python library, we define these expression variables as one of many kinds of Theano variable types.
```
x = T.dscalar()
```
So dscalar() is a type of Theano variable or data type that is computationally represented as a float64. There are many other data types available (see <a href="http://deeplearning.net/software/theano/library/tensor/basic.html">here</a>), but we're interested in just defining a single variable that is a scalar.
Now let's build out the expression.
```
fx = T.exp(T.sin(x**2))
```
Here I've defined our expression that is equivalent to the mathematical one above. `fx` is now a variable itself that depends on the `x` variable.
```
type(fx) #just to show you that fx is a theano variable type
```
Okay, so that's nice. What now? Well, now we need to "compile" this expression into a Theano function. Theano will do some magic behind the scenes including building a computational graph, optimizing operations, and compiling to C code to get this to run fast and allow it to compute gradients.
```
f = theano.function(inputs=[x], outputs=[fx])
f(10)
```
We compiled our `fx` expression into a Theano function. As you can see, `theano.function` has two required arguments, inputs and outputs. Our only input is our Theano variable `x` and our output is our `fx` expression. Then we ran the f() function supplying it with the value `10` and it accurately spit out the computation. So up until this point we could have easily just `np.exp(np.sin(100))` using numpy and get the same result. But that would be an exact, imperative, computation and not a symbolic computational graph. Now let's show off Theano's autodifferentiation.
To do that, we'll use `T.grad()` which will give us a symbolically differentiated expression of our function, then we pass it to `theano.function` to compile a new function to call it. `wrt` stands for 'with respect to', i.e. we're deriving our expression `fx` with respect to it's variable `x`.
```
fp = T.grad(fx, wrt=x)
fprime = theano.function([x], fp)
fprime(15)
```
4.347 is indeed the derivative of our expression evaluated at $x=15$, don't worry, I checked with WolframAlpha. And to be clear, Theano can take the derivative of arbitrarily complex expressions. Don't be fooled by our extremely simple starter expression here. Automatically calculating gradients is a huge help since it saves us the time of having to manually come up with the gradient expressions for whatever neural network we build.
So there you have it. Those are the very basics of Theano. We're going to utilize a few other features of Theano in the neural net we'll build but not much.
####Now, for an XOR neural network
We're going to symbolically define two Theano variables called `x` and `y`. We're going to build our familiar XOR network with 2 input units (+ a bias), 2 hidden units (+ a bias), and 1 output unit. So our `x` variable will always be a 2-element vector (e.g. [0,1]) and our `y` variable will always be a scalar and is our expected value for each pair of `x` values.
```
x = T.dvector()
y = T.dscalar()
```
Now let's define a Python function that will be a matrix multiplier and sigmoid function, so it will accept and `x` vector (and concatenate in a bias value of 1) and a `w` weight matrix, multiply them, and then run them through a sigmoid function. Theano has the sigmoid function built in the `nnet` class that we imported above. We'll use this function as our basic layer output function.
```
def layer(x, w):
b = np.array([1], dtype=theano.config.floatX)
new_x = T.concatenate([x, b])
m = T.dot(w.T, new_x) #theta1: 3x3 * x: 3x1 = 3x1 ;;; theta2: 1x4 * 4x1
h = nnet.sigmoid(m)
return h
```
Theano can be a bit touchy. In order to concatenate a scalar value of 1 to our 1-dimensional vector `x`, we create a numpy array with a single element (`1`), and explicitly pass in the `dtype` parameter to make it a float64 and compatible with our Theano vector variable. You'll also notice that Theano provides its own version of many numpy functions, such as the dot product that we're using. Theano can work with numpy but in the end it all has to get converted to Theano types.
This feels a little bit premature, but let's go ahead and implement our gradient descent function. Don't worry, it's very simple. We're just going to have a function that defines a learning rate `alpha` and accepts a cost/error expression and a weight matrix. It will use Theano's `grad()` function to compute the gradient of the cost function with respect to the given weight matrix and return an updated weight matrix.
```
def grad_desc(cost, theta):
alpha = 0.1 #learning rate
return theta - (alpha * T.grad(cost, wrt=theta))
```
We're making good progress. At this point we can define our weight matrices and initialize them to random values.
Since our weight matrices will take on definite values, they're not going to be represented as Theano variables, they're going to be defined as Theano's _shared_ variable. A shared variable is what we use for things we want to give a definite value but we also want to update. Notice that I didn't define the `alpha` or `b` (the bias term) as shared variables, I just hard-coded them as strict values because I am never going to update/modify them.
```
theta1 = theano.shared(np.array(np.random.rand(3,3), dtype=theano.config.floatX)) # randomly initialize
theta2 = theano.shared(np.array(np.random.rand(4,1), dtype=theano.config.floatX))
```
So here we've defined our two weight matrices for our 3 layer network and initialized them using numpy's random class. Again we specifically define the dtype parameter so it will be a float64, compatible with our Theano `dscalar` and `dvector` variable types.
Here's where the fun begins. We can start actually doing our computations for each layer in the network. Of course we'll start by computing the hidden layer's output using our previously defined `layer` function, and pass in the Theano `x` variable we defined above and our `theta1` matrix.
```
hid1 = layer(x, theta1) #hidden layer
```
We can do the same for our final output layer. Notice I use the T.sum() function on the outside which is the same as numpy's sum(). This is only because Theano will complain if you don't make it explicitly clear that our output is returning a scalar and not a matrix. Our matrix dimensional analysis is sure to return a 1x1 single element vector but we need to convert it to a scalar since we're substracting `out1` from `y` in our cost expression that follows.
```
out1 = T.sum(layer(hid1, theta2)) #output layer
fc = (out1 - y)**2 #cost expression
```
Ahh, almost done. We're going to compile two Theano functions. One will be our cost expression (for training), and the other will be our output layer expression (to run the network forward).
```
cost = theano.function(inputs=[x, y], outputs=fc, updates=[
(theta1, grad_desc(fc, theta1)),
(theta2, grad_desc(fc, theta2))])
run_forward = theano.function(inputs=[x], outputs=out1)
```
Our `theano.function` call looks a bit different than in our first example. Yeah, we have this additional `updates` parameter. `updates` allows us to update our shared variables according to an expression. `updates` expects a list of 2-tuples:
```python
updates=[(shared_variable, update_value), ...]
```
The second part of each tuple can be an expression or function that returns the new value we want to update the first part to. In our case, we have two shared variables we want to update, `theta1` and `theta2` and we want to use our `grad_desc` function to give us the updated data. Of course our `grad_desc` function expects two arguments, a cost function and a weight matrix, so we pass those in. `fc` is our cost expression. So every time we invoke/call the `cost` function that we've compiled with Theano, it will also update our shared variables according to our `grad_desc` rule. Pretty convenient!
Additionally, we've compiled a `run_forward` function just so we can run the network forward and make sure it has trained properly. We don't need to update anything there.
Now let's define our training data and setup a `for` loop to iterate through our training epochs.
```
inputs = np.array([[0,1],[1,0],[1,1],[0,0]]).reshape(4,2) #training data X
exp_y = np.array([1, 1, 0, 0]) #training data Y
cur_cost = 0
for i in range(10000):
for k in range(len(inputs)):
cur_cost = cost(inputs[k], exp_y[k]) #call our Theano-compiled cost function, it will auto update weights
if i % 500 == 0: #only print the cost every 500 epochs/iterations (to save space)
print('Cost: %s' % (cur_cost,))
#Training done! Let's test it out
print(run_forward([0,1]))
print(run_forward([1,1]))
print(run_forward([1,0]))
print(run_forward([0,0]))
```
It works!
####Closing words
Theano is a pretty robust and complicated library but hopefully this simple introduction helps you get started. I certainly struggled with it before it made sense to me. And clearly using Theano for an XOR neural network is overkill, but its optimization power and GPU utilization really comes into play for bigger projects. Nonetheless, not having to think about manually calculating gradients is nice.
Cheers
####References:
1. http://deeplearning.net/software/theano/index.html
2. https://gist.github.com/honnibal/6a9e5ef2921c0214eeeb
|
github_jupyter
|
import theano
import theano.tensor as T
import theano.tensor.nnet as nnet
import numpy as np
x = T.dscalar()
fx = T.exp(T.sin(x**2))
type(fx) #just to show you that fx is a theano variable type
f = theano.function(inputs=[x], outputs=[fx])
f(10)
fp = T.grad(fx, wrt=x)
fprime = theano.function([x], fp)
fprime(15)
x = T.dvector()
y = T.dscalar()
def layer(x, w):
b = np.array([1], dtype=theano.config.floatX)
new_x = T.concatenate([x, b])
m = T.dot(w.T, new_x) #theta1: 3x3 * x: 3x1 = 3x1 ;;; theta2: 1x4 * 4x1
h = nnet.sigmoid(m)
return h
def grad_desc(cost, theta):
alpha = 0.1 #learning rate
return theta - (alpha * T.grad(cost, wrt=theta))
theta1 = theano.shared(np.array(np.random.rand(3,3), dtype=theano.config.floatX)) # randomly initialize
theta2 = theano.shared(np.array(np.random.rand(4,1), dtype=theano.config.floatX))
hid1 = layer(x, theta1) #hidden layer
out1 = T.sum(layer(hid1, theta2)) #output layer
fc = (out1 - y)**2 #cost expression
cost = theano.function(inputs=[x, y], outputs=fc, updates=[
(theta1, grad_desc(fc, theta1)),
(theta2, grad_desc(fc, theta2))])
run_forward = theano.function(inputs=[x], outputs=out1)
updates=[(shared_variable, update_value), ...]
inputs = np.array([[0,1],[1,0],[1,1],[0,0]]).reshape(4,2) #training data X
exp_y = np.array([1, 1, 0, 0]) #training data Y
cur_cost = 0
for i in range(10000):
for k in range(len(inputs)):
cur_cost = cost(inputs[k], exp_y[k]) #call our Theano-compiled cost function, it will auto update weights
if i % 500 == 0: #only print the cost every 500 epochs/iterations (to save space)
print('Cost: %s' % (cur_cost,))
#Training done! Let's test it out
print(run_forward([0,1]))
print(run_forward([1,1]))
print(run_forward([1,0]))
print(run_forward([0,0]))
| 0.325413 | 0.98537 |
<div align="center">
<h1><img width="30" src="https://madewithml.com/static/images/rounded_logo.png"> <a href="https://madewithml.com/">Made With ML</a></h1>
Applied ML · MLOps · Production
<br>
Join 30K+ developers in learning how to responsibly <a href="https://madewithml.com/about/">deliver value</a> with ML.
<br>
</div>
<br>
<div align="center">
<a target="_blank" href="https://newsletter.madewithml.com"><img src="https://img.shields.io/badge/Subscribe-30K-brightgreen"></a>
<a target="_blank" href="https://github.com/GokuMohandas/MadeWithML"><img src="https://img.shields.io/github/stars/GokuMohandas/MadeWithML.svg?style=social&label=Star"></a>
<a target="_blank" href="https://www.linkedin.com/in/goku"><img src="https://img.shields.io/badge/style--5eba00.svg?label=LinkedIn&logo=linkedin&style=social"></a>
<a target="_blank" href="https://twitter.com/GokuMohandas"><img src="https://img.shields.io/twitter/follow/GokuMohandas.svg?label=Follow&style=social"></a>
<br>
🔥 Among the <a href="https://github.com/topics/deep-learning" target="_blank">top ML</a> repositories on GitHub
</div>
<br>
<hr>
# Notebooks
In this lesson, we'll learn about how to work with notebooks.
<div align="left">
<a target="_blank" href="https://madewithml.com/courses/basics/notebooks/"><img src="https://img.shields.io/badge/📖 Read-blog post-9cf"></a>
<a href="https://github.com/GokuMohandas/MadeWithML/blob/main/notebooks/01_Notebooks.ipynb" role="button"><img src="https://img.shields.io/static/v1?label=&message=View%20On%20GitHub&color=586069&logo=github&labelColor=2f363d"></a>
<a href="https://colab.research.google.com/github/GokuMohandas/MadeWithML/blob/main/notebooks/01_Notebooks.ipynb"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"></a>
</div>
# Set Up
1. Click on this link to open the accompanying [notebook]() for this lesson or create a blank one on [Google Colab](https://colab.research.google.com/).
2. Sign into your [Google account](https://accounts.google.com/signin) to start using the notebook. If you don't want to save your work, you can skip the steps below. If you do not have access to Google, you can follow along using [Jupyter Lab](https://jupyter.org/).
3. If you do want to save your work, click the **COPY TO DRIVE** button on the toolbar. This will open a new notebook in a new tab. Rename this new notebook by removing the words Copy of from the title (change `Copy of 01_Notebooks` to `01_Notebooks`).
<div align="center">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/notebooks/copy_to_drive.png" width="400">  <img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/notebooks/rename.png" width="320">
</div>
# Types of cells
Notebooks are made up of cells. Each cell can either be a `code cell` or a `text cell`.
* `code cell`: used for writing and executing code.
* `text cell`: used for writing text, HTML, Markdown, etc.
# Creating cells
First, let's create a text cell. Click on a desired location in the notebook and create the cell by clicking on the `➕ TEXT` (located in the top left corner).
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/notebooks/text_cell.png" width="320">
</div>
Once you create the cell, click on it and type the following text inside it:
```
### This is a header
Hello world!
```
### This is a header
Hello world!
# Running cells
Once you type inside the cell, press the `SHIFT` and `RETURN` (enter key) together to run the cell.
# Editing cells
To edit a cell, double click on it and make any changes.
# Moving cells
Once you create the cell, you can move it up and down by clicking on the cell and then pressing the ⬆ and ⬇ button on the top right of the cell.
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/notebooks/move_cell.png" width="500">
</div>
# Deleting cells
You can delete the cell by clicking on it and pressing the trash can button 🗑️ on the top right corner of the cell. Alternatively, you can also press ⌘/Ctrl + M + D.
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/notebooks/delete_cell.png" width="500">
</div>
# Creating a code cell
You can repeat the steps above to create and edit a *code* cell. You can create a code cell by clicking on the `➕ CODE` (located in the top left corner).
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/notebooks/code_cell.png" width="320">
</div>
Once you've created the code cell, double click on it, type the following inside it and then press `Shift + Enter` to execute the code.
```
print ("Hello world!")
```
```
print ("Hello world!")
```
These are the basic concepts you'll need to use these notebooks but we'll learn few more tricks in subsequent lessons.
|
github_jupyter
|
### This is a header
Hello world!
print ("Hello world!")
print ("Hello world!")
| 0.31363 | 0.969814 |
```
import numpy as np
import pulp
# create the LP object, set up as a maximization problem
prob = pulp.LpProblem('Giapetto', pulp.LpMaximize)
# set up decision variables
soldiers = pulp.LpVariable('soldiers', lowBound=0, cat='Integer')
trains = pulp.LpVariable('trains', lowBound=0, cat='Integer')
# model weekly production costs
raw_material_costs = 10 * soldiers + 9 * trains
variable_costs = 14 * soldiers + 10 * trains
# model weekly revenues from toy sales
revenues = 27 * soldiers + 21 * trains
# use weekly profit as the objective function to maximize
profit = revenues - (raw_material_costs + variable_costs)
prob += profit # here's where we actually add it to the obj function
# add constraints for available labor hours
carpentry_hours = soldiers + trains
prob += (carpentry_hours <= 80)
finishing_hours = 2*soldiers + trains
prob += (finishing_hours <= 100)
# add constraint representing demand for soldiers
prob += (soldiers <= 40)
# solve the LP using the default solver
optimization_result = prob.solve()
# make sure we got an optimal solution
assert optimization_result == pulp.LpStatusOptimal
# display the results
for var in (soldiers, trains):
print('Optimal weekly number of {} to produce: {:1.0f}'.format(var.name, var.value()))
from matplotlib import pyplot as plt
from matplotlib.path import Path
from matplotlib.patches import PathPatch
# use seaborn to change the default graphics to something nicer
# and set a nice color palette
import seaborn as sns
sns.set_palette('Set1')
# create the plot object
fig, ax = plt.subplots(figsize=(8, 8))
s = np.linspace(0, 100)
# add carpentry constraint: trains <= 80 - soldiers
plt.plot(s, 80 - s, lw=3, label='carpentry')
plt.fill_between(s, 0, 80 - s, alpha=0.1)
# add finishing constraint: trains <= 100 - 2*soldiers
plt.plot(s, 100 - 2 * s, lw=3, label='finishing')
plt.fill_between(s, 0, 100 - 2 * s, alpha=0.1)
# add demains constraint: soldiers <= 40
plt.plot(40 * np.ones_like(s), s, lw=3, label='demand')
plt.fill_betweenx(s, 0, 40, alpha=0.1)
# add non-negativity constraints
plt.plot(np.zeros_like(s), s, lw=3, label='t non-negative')
plt.plot(s, np.zeros_like(s), lw=3, label='s non-negative')
# highlight the feasible region
path = Path([
(0., 0.),
(0., 80.),
(20., 60.),
(40., 20.),
(40., 0.),
(0., 0.),
])
patch = PathPatch(path, label='feasible region', alpha=0.5)
ax.add_patch(patch)
# labels and stuff
plt.xlabel('soldiers', fontsize=16)
plt.ylabel('trains', fontsize=16)
plt.xlim(-0.5, 100)
plt.ylim(-0.5, 100)
plt.legend(fontsize=14)
plt.show()
```
|
github_jupyter
|
import numpy as np
import pulp
# create the LP object, set up as a maximization problem
prob = pulp.LpProblem('Giapetto', pulp.LpMaximize)
# set up decision variables
soldiers = pulp.LpVariable('soldiers', lowBound=0, cat='Integer')
trains = pulp.LpVariable('trains', lowBound=0, cat='Integer')
# model weekly production costs
raw_material_costs = 10 * soldiers + 9 * trains
variable_costs = 14 * soldiers + 10 * trains
# model weekly revenues from toy sales
revenues = 27 * soldiers + 21 * trains
# use weekly profit as the objective function to maximize
profit = revenues - (raw_material_costs + variable_costs)
prob += profit # here's where we actually add it to the obj function
# add constraints for available labor hours
carpentry_hours = soldiers + trains
prob += (carpentry_hours <= 80)
finishing_hours = 2*soldiers + trains
prob += (finishing_hours <= 100)
# add constraint representing demand for soldiers
prob += (soldiers <= 40)
# solve the LP using the default solver
optimization_result = prob.solve()
# make sure we got an optimal solution
assert optimization_result == pulp.LpStatusOptimal
# display the results
for var in (soldiers, trains):
print('Optimal weekly number of {} to produce: {:1.0f}'.format(var.name, var.value()))
from matplotlib import pyplot as plt
from matplotlib.path import Path
from matplotlib.patches import PathPatch
# use seaborn to change the default graphics to something nicer
# and set a nice color palette
import seaborn as sns
sns.set_palette('Set1')
# create the plot object
fig, ax = plt.subplots(figsize=(8, 8))
s = np.linspace(0, 100)
# add carpentry constraint: trains <= 80 - soldiers
plt.plot(s, 80 - s, lw=3, label='carpentry')
plt.fill_between(s, 0, 80 - s, alpha=0.1)
# add finishing constraint: trains <= 100 - 2*soldiers
plt.plot(s, 100 - 2 * s, lw=3, label='finishing')
plt.fill_between(s, 0, 100 - 2 * s, alpha=0.1)
# add demains constraint: soldiers <= 40
plt.plot(40 * np.ones_like(s), s, lw=3, label='demand')
plt.fill_betweenx(s, 0, 40, alpha=0.1)
# add non-negativity constraints
plt.plot(np.zeros_like(s), s, lw=3, label='t non-negative')
plt.plot(s, np.zeros_like(s), lw=3, label='s non-negative')
# highlight the feasible region
path = Path([
(0., 0.),
(0., 80.),
(20., 60.),
(40., 20.),
(40., 0.),
(0., 0.),
])
patch = PathPatch(path, label='feasible region', alpha=0.5)
ax.add_patch(patch)
# labels and stuff
plt.xlabel('soldiers', fontsize=16)
plt.ylabel('trains', fontsize=16)
plt.xlim(-0.5, 100)
plt.ylim(-0.5, 100)
plt.legend(fontsize=14)
plt.show()
| 0.504883 | 0.555375 |
# A Simple Example of usage
In this example I will go through creating the following entities in order:
- warehouse
- shipping zone
- product attributes with their values.
- categories
- product type
- products with variants and their stocks.
I will use Tea product as a fictive example.
```
from saleor_gql_loader import ETLDataLoader
# I generated a token for my app as explained in the README.md
# https://github.com/grll/saleor-gql-loader/blob/master/README.md
etl_data_loader = ETLDataLoader("LcLNVgUt8mu8yKJ0Wrh3nADnTT21uv")
# create a default warehouse
warehouse_id = etl_data_loader.create_warehouse()
warehouse_id
# create a default shipping zone associated
shipping_zone_id = etl_data_loader.create_shipping_zone(addWarehouses=[warehouse_id])
shipping_zone_id
# define my products usually extracted from csv or scraped...
products = [
{
"name": "tea a",
"description": "description for tea a",
"category": "green tea",
"price": 5.5,
"strength": "medium"
},
{
"name": "tea b",
"description": "description for tea b",
"category": "black tea",
"price": 10.5,
"strength": "strong"
},
{
"name": "tea c",
"description": "description for tea c",
"category": "green tea",
"price": 9.5,
"strength": "light"
}
]
# add basic sku to products
for i, product in enumerate(products):
product["sku"] = "{:05}-00".format(i)
# create the strength attribute
strength_attribute_id = etl_data_loader.create_attribute(name="strength")
unique_strength = set([product['strength'] for product in products])
for strength in unique_strength:
etl_data_loader.create_attribute_value(strength_attribute_id, name=strength)
# create another quantity attribute used as variant:
qty_attribute_id = etl_data_loader.create_attribute(name="qty")
unique_qty = {"100g", "200g", "300g"}
for qty in unique_qty:
etl_data_loader.create_attribute_value(qty_attribute_id, name=qty)
# create a product type: tea
product_type_id = etl_data_loader.create_product_type(name="tea",
hasVariants=True,
productAttributes=[strength_attribute_id],
variantAttributes=[qty_attribute_id])
# create categories
unique_categories = set([product['category'] for product in products])
cat_to_id = {}
for category in unique_categories:
cat_to_id[category] = etl_data_loader.create_category(name=category)
cat_to_id
# create products and store id
for i, product in enumerate(products):
product_id = etl_data_loader.create_product(product_type_id,
name=product["name"],
description=product["description"],
basePrice=product["price"],
sku=product["sku"],
category=cat_to_id[product["category"]],
attributes=[{"id": strength_attribute_id, "values": [product["strength"]]}],
isPublished=True)
products[i]["id"] = product_id
# create some variant for each product:
for product in products:
for i, qty in enumerate(unique_qty):
variant_id = etl_data_loader.create_product_variant(product_id,
sku=product["sku"].replace("-00", "-1{}".format(i+1)),
attributes=[{"id": qty_attribute_id, "values": [qty]}],
costPrice=product["price"],
weight=0.75,
stocks=[{"warehouse": warehouse_id, "quantity": 15}])
```
|
github_jupyter
|
from saleor_gql_loader import ETLDataLoader
# I generated a token for my app as explained in the README.md
# https://github.com/grll/saleor-gql-loader/blob/master/README.md
etl_data_loader = ETLDataLoader("LcLNVgUt8mu8yKJ0Wrh3nADnTT21uv")
# create a default warehouse
warehouse_id = etl_data_loader.create_warehouse()
warehouse_id
# create a default shipping zone associated
shipping_zone_id = etl_data_loader.create_shipping_zone(addWarehouses=[warehouse_id])
shipping_zone_id
# define my products usually extracted from csv or scraped...
products = [
{
"name": "tea a",
"description": "description for tea a",
"category": "green tea",
"price": 5.5,
"strength": "medium"
},
{
"name": "tea b",
"description": "description for tea b",
"category": "black tea",
"price": 10.5,
"strength": "strong"
},
{
"name": "tea c",
"description": "description for tea c",
"category": "green tea",
"price": 9.5,
"strength": "light"
}
]
# add basic sku to products
for i, product in enumerate(products):
product["sku"] = "{:05}-00".format(i)
# create the strength attribute
strength_attribute_id = etl_data_loader.create_attribute(name="strength")
unique_strength = set([product['strength'] for product in products])
for strength in unique_strength:
etl_data_loader.create_attribute_value(strength_attribute_id, name=strength)
# create another quantity attribute used as variant:
qty_attribute_id = etl_data_loader.create_attribute(name="qty")
unique_qty = {"100g", "200g", "300g"}
for qty in unique_qty:
etl_data_loader.create_attribute_value(qty_attribute_id, name=qty)
# create a product type: tea
product_type_id = etl_data_loader.create_product_type(name="tea",
hasVariants=True,
productAttributes=[strength_attribute_id],
variantAttributes=[qty_attribute_id])
# create categories
unique_categories = set([product['category'] for product in products])
cat_to_id = {}
for category in unique_categories:
cat_to_id[category] = etl_data_loader.create_category(name=category)
cat_to_id
# create products and store id
for i, product in enumerate(products):
product_id = etl_data_loader.create_product(product_type_id,
name=product["name"],
description=product["description"],
basePrice=product["price"],
sku=product["sku"],
category=cat_to_id[product["category"]],
attributes=[{"id": strength_attribute_id, "values": [product["strength"]]}],
isPublished=True)
products[i]["id"] = product_id
# create some variant for each product:
for product in products:
for i, qty in enumerate(unique_qty):
variant_id = etl_data_loader.create_product_variant(product_id,
sku=product["sku"].replace("-00", "-1{}".format(i+1)),
attributes=[{"id": qty_attribute_id, "values": [qty]}],
costPrice=product["price"],
weight=0.75,
stocks=[{"warehouse": warehouse_id, "quantity": 15}])
| 0.737064 | 0.747455 |
# Machine learning Grid Search
## Imports
```
import sys
import cufflinks
import pandas as pd
import numpy as np
from tqdm import tqdm
import warnings
import pickle
warnings.filterwarnings('ignore')
sys.path.append('./..')
cufflinks.go_offline()
from Corpus.Corpus import get_corpus, filter_binary_pn, filter_corpus_small
from auxiliar.VectorizerHelper import vectorizer, vectorizerIdf, preprocessor
from auxiliar import parameters
from auxiliar.HtmlParser import HtmlParser
from sklearn.calibration import CalibratedClassifierCV
from sklearn.ensemble import RandomForestClassifier
from sklearn.multiclass import OneVsRestClassifier
from sklearn.svm import LinearSVC
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import MultinomialNB
from sklearn.model_selection import GridSearchCV
from sklearn.pipeline import Pipeline
from sklearn.metrics import accuracy_score
from sklearn.metrics import mean_squared_error
from sklearn.metrics import roc_auc_score
from sklearn.metrics import f1_score
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from sklearn.metrics import precision_score
from sklearn.metrics import roc_curve
from sklearn.model_selection import KFold
import copy
```
## Config
```
polarity_dim = 5
clasificadores=['lr', 'ls', 'mb', 'rf']
idf = False
target_names=['Neg', 'Pos']
kfolds = 10
base_dir = '2-clases' if polarity_dim == 2 else ('3-clases' if polarity_dim == 3 else '5-clases')
name = 'machine_learning/tweeter/grid_search'
```
## Get data
```
cine = HtmlParser(200, "http://www.muchocine.net/criticas_ultimas.php", 1)
data_corpus = get_corpus('general-corpus', 'general-corpus', 1, None)
if polarity_dim == 2:
data_corpus = filter_binary_pn(data_corpus)
cine = filter_binary_pn(cine.get_corpus())
elif polarity_dim == 3:
data_corpus = filter_corpus_small(data_corpus)
cine = filter_corpus_small(cine.get_corpus())
elif polarity_dim == 5:
cine = cine.get_corpus()
cine = cine[:5000]
used_data = pd.DataFrame(data_corpus)
```
## Split data
```
split = used_data.shape[0] * 0.7
train_corpus = used_data.loc[:split - 1 , :]
test_corpus = used_data.loc[split:, :]
```
## Initialize ML
```
vect = vectorizerIdf if idf else vectorizer
ls = CalibratedClassifierCV(LinearSVC()) if polarity_dim == 2 else OneVsRestClassifier(CalibratedClassifierCV(LinearSVC()))
lr = LogisticRegression(solver='lbfgs') if polarity_dim == 2 else OneVsRestClassifier(LogisticRegression())
mb = MultinomialNB() if polarity_dim == 2 else OneVsRestClassifier(MultinomialNB())
rf = RandomForestClassifier() if polarity_dim == 2 else OneVsRestClassifier(RandomForestClassifier())
pipeline_ls = Pipeline([
('prep', copy.deepcopy(preprocessor)),
('vect', copy.deepcopy(vect)),
('ls', ls)
])
pipeline_lr = Pipeline([
('prep', copy.deepcopy(preprocessor)),
('vect', copy.deepcopy(vect)),
('lr', lr)
])
pipeline_mb = Pipeline([
('prep', copy.deepcopy(preprocessor)),
('vect', copy.deepcopy(vect)),
('mb', mb)
])
pipeline_rf = Pipeline([
('prep', copy.deepcopy(preprocessor)),
('vect', copy.deepcopy(vect)),
('rf', rf)
])
pipelines = {
'ls': pipeline_ls,
'lr': pipeline_lr,
'mb': pipeline_mb,
'rf': pipeline_rf
}
pipelines_train = {
'ls': ls,
'lr': lr,
'mb': mb,
'rf': rf
}
params = parameters.parameters_bin if polarity_dim == 2 else parameters.parameters
params
```
## Train
```
folds = pd.read_pickle('./../data/pkls/folds.pkl') # k-folds precargados
folds = folds.values
pd.Series(train_corpus.content.values)
results = {}
grids = {}
with tqdm(total=len(clasificadores) * 10) as pbar:
for c in clasificadores:
results[c] = { 'real': {}, 'predicted': {} }
i = 0
params[c].update(parameters.vect_params)
params[c].update(parameters.prepro_params)
param_grid = params[c]
grid_search = GridSearchCV(pipelines[c], param_grid, verbose=2, scoring='accuracy', refit=False, cv=3)
grid = grid_search.fit(train_corpus.content, train_corpus.polarity)
grids[c] = grid
best_parameters = grid.best_params_
train_params = {}
for param_name in sorted(parameters.vect_params.keys()):
train_params.update({param_name[6:]: best_parameters[param_name]})
vect.set_params(**train_params)
preprocessor.set_params(**train_params)
x_prepro = preprocessor.fit_transform(train_corpus.content)
x_vect = vect.fit_transform(x_prepro, train_corpus.polarity).toarray()
for train_index, test_index in folds:
train_x = x_vect[train_index]
train_y = train_corpus.polarity[train_index]
test_x = x_vect[test_index]
test_y = train_corpus.polarity[test_index]
pipelines_train[c].fit(train_x, train_y)
predicted = pipelines_train[c].predict(test_x)
results[c]['real'][i] = test_y.values.tolist()
results[c]['predicted'][i] = predicted.tolist()
i = i + 1
pbar.update(1)
results
pd.DataFrame(results).to_pickle('../results/'+name+'/'+base_dir+'/results.pkl')
with open('../results/'+name+'/'+base_dir+'/grid_results.pkl', 'wb') as fp:
pickle.dump(grids, fp)
with open('../results/'+name+'/'+base_dir+'/grid_results-idf.pkl', 'rb') as fp:
grids = pickle.load(fp)
test_results = {}
with tqdm(total=len(clasificadores)) as pbar:
for c in clasificadores:
test_results[c] = { 'real': {}, 'cine_real': {}, 'predicted': {}, 'cine_predicted': {} }
i = 0
grid = grids[c]
best_parameters = grid.best_params_
train_params = {}
for param_name in sorted(parameters.vect_params.keys()):
train_params.update({param_name[6:]: best_parameters[param_name]})
vect.set_params(**train_params)
vect.fit(data_corpus.content, data_corpus.polarity)
x_vect = vect.transform(train_corpus.content).toarray()
x_vect_test = vect.transform(test_corpus.content).toarray()
x_vect_cine = vect.transform(cine.content).toarray()
train_x = x_vect
train_y = train_corpus.polarity
test_x = x_vect_test
test_y = test_corpus.polarity
cine_y = cine.polarity
pipelines_train[c].fit(train_x, train_y)
predicted = pipelines_train[c].predict(test_x)
cine_predicted = pipelines_train[c].predict(x_vect_cine)
test_results[c]['real'][i] = test_y.values.tolist()
test_results[c]['cine_real'][i] = cine_y.values.tolist()
test_results[c]['predicted'][i] = predicted.tolist()
test_results[c]['cine_predicted'][i] = cine_predicted.tolist()
i = i + 1
pbar.update(1)
pbar.update(1)
pd.DataFrame(test_results).to_pickle('../results/'+name+'/'+base_dir+'/test_results.pkl')
```
|
github_jupyter
|
import sys
import cufflinks
import pandas as pd
import numpy as np
from tqdm import tqdm
import warnings
import pickle
warnings.filterwarnings('ignore')
sys.path.append('./..')
cufflinks.go_offline()
from Corpus.Corpus import get_corpus, filter_binary_pn, filter_corpus_small
from auxiliar.VectorizerHelper import vectorizer, vectorizerIdf, preprocessor
from auxiliar import parameters
from auxiliar.HtmlParser import HtmlParser
from sklearn.calibration import CalibratedClassifierCV
from sklearn.ensemble import RandomForestClassifier
from sklearn.multiclass import OneVsRestClassifier
from sklearn.svm import LinearSVC
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import MultinomialNB
from sklearn.model_selection import GridSearchCV
from sklearn.pipeline import Pipeline
from sklearn.metrics import accuracy_score
from sklearn.metrics import mean_squared_error
from sklearn.metrics import roc_auc_score
from sklearn.metrics import f1_score
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from sklearn.metrics import precision_score
from sklearn.metrics import roc_curve
from sklearn.model_selection import KFold
import copy
polarity_dim = 5
clasificadores=['lr', 'ls', 'mb', 'rf']
idf = False
target_names=['Neg', 'Pos']
kfolds = 10
base_dir = '2-clases' if polarity_dim == 2 else ('3-clases' if polarity_dim == 3 else '5-clases')
name = 'machine_learning/tweeter/grid_search'
cine = HtmlParser(200, "http://www.muchocine.net/criticas_ultimas.php", 1)
data_corpus = get_corpus('general-corpus', 'general-corpus', 1, None)
if polarity_dim == 2:
data_corpus = filter_binary_pn(data_corpus)
cine = filter_binary_pn(cine.get_corpus())
elif polarity_dim == 3:
data_corpus = filter_corpus_small(data_corpus)
cine = filter_corpus_small(cine.get_corpus())
elif polarity_dim == 5:
cine = cine.get_corpus()
cine = cine[:5000]
used_data = pd.DataFrame(data_corpus)
split = used_data.shape[0] * 0.7
train_corpus = used_data.loc[:split - 1 , :]
test_corpus = used_data.loc[split:, :]
vect = vectorizerIdf if idf else vectorizer
ls = CalibratedClassifierCV(LinearSVC()) if polarity_dim == 2 else OneVsRestClassifier(CalibratedClassifierCV(LinearSVC()))
lr = LogisticRegression(solver='lbfgs') if polarity_dim == 2 else OneVsRestClassifier(LogisticRegression())
mb = MultinomialNB() if polarity_dim == 2 else OneVsRestClassifier(MultinomialNB())
rf = RandomForestClassifier() if polarity_dim == 2 else OneVsRestClassifier(RandomForestClassifier())
pipeline_ls = Pipeline([
('prep', copy.deepcopy(preprocessor)),
('vect', copy.deepcopy(vect)),
('ls', ls)
])
pipeline_lr = Pipeline([
('prep', copy.deepcopy(preprocessor)),
('vect', copy.deepcopy(vect)),
('lr', lr)
])
pipeline_mb = Pipeline([
('prep', copy.deepcopy(preprocessor)),
('vect', copy.deepcopy(vect)),
('mb', mb)
])
pipeline_rf = Pipeline([
('prep', copy.deepcopy(preprocessor)),
('vect', copy.deepcopy(vect)),
('rf', rf)
])
pipelines = {
'ls': pipeline_ls,
'lr': pipeline_lr,
'mb': pipeline_mb,
'rf': pipeline_rf
}
pipelines_train = {
'ls': ls,
'lr': lr,
'mb': mb,
'rf': rf
}
params = parameters.parameters_bin if polarity_dim == 2 else parameters.parameters
params
folds = pd.read_pickle('./../data/pkls/folds.pkl') # k-folds precargados
folds = folds.values
pd.Series(train_corpus.content.values)
results = {}
grids = {}
with tqdm(total=len(clasificadores) * 10) as pbar:
for c in clasificadores:
results[c] = { 'real': {}, 'predicted': {} }
i = 0
params[c].update(parameters.vect_params)
params[c].update(parameters.prepro_params)
param_grid = params[c]
grid_search = GridSearchCV(pipelines[c], param_grid, verbose=2, scoring='accuracy', refit=False, cv=3)
grid = grid_search.fit(train_corpus.content, train_corpus.polarity)
grids[c] = grid
best_parameters = grid.best_params_
train_params = {}
for param_name in sorted(parameters.vect_params.keys()):
train_params.update({param_name[6:]: best_parameters[param_name]})
vect.set_params(**train_params)
preprocessor.set_params(**train_params)
x_prepro = preprocessor.fit_transform(train_corpus.content)
x_vect = vect.fit_transform(x_prepro, train_corpus.polarity).toarray()
for train_index, test_index in folds:
train_x = x_vect[train_index]
train_y = train_corpus.polarity[train_index]
test_x = x_vect[test_index]
test_y = train_corpus.polarity[test_index]
pipelines_train[c].fit(train_x, train_y)
predicted = pipelines_train[c].predict(test_x)
results[c]['real'][i] = test_y.values.tolist()
results[c]['predicted'][i] = predicted.tolist()
i = i + 1
pbar.update(1)
results
pd.DataFrame(results).to_pickle('../results/'+name+'/'+base_dir+'/results.pkl')
with open('../results/'+name+'/'+base_dir+'/grid_results.pkl', 'wb') as fp:
pickle.dump(grids, fp)
with open('../results/'+name+'/'+base_dir+'/grid_results-idf.pkl', 'rb') as fp:
grids = pickle.load(fp)
test_results = {}
with tqdm(total=len(clasificadores)) as pbar:
for c in clasificadores:
test_results[c] = { 'real': {}, 'cine_real': {}, 'predicted': {}, 'cine_predicted': {} }
i = 0
grid = grids[c]
best_parameters = grid.best_params_
train_params = {}
for param_name in sorted(parameters.vect_params.keys()):
train_params.update({param_name[6:]: best_parameters[param_name]})
vect.set_params(**train_params)
vect.fit(data_corpus.content, data_corpus.polarity)
x_vect = vect.transform(train_corpus.content).toarray()
x_vect_test = vect.transform(test_corpus.content).toarray()
x_vect_cine = vect.transform(cine.content).toarray()
train_x = x_vect
train_y = train_corpus.polarity
test_x = x_vect_test
test_y = test_corpus.polarity
cine_y = cine.polarity
pipelines_train[c].fit(train_x, train_y)
predicted = pipelines_train[c].predict(test_x)
cine_predicted = pipelines_train[c].predict(x_vect_cine)
test_results[c]['real'][i] = test_y.values.tolist()
test_results[c]['cine_real'][i] = cine_y.values.tolist()
test_results[c]['predicted'][i] = predicted.tolist()
test_results[c]['cine_predicted'][i] = cine_predicted.tolist()
i = i + 1
pbar.update(1)
pbar.update(1)
pd.DataFrame(test_results).to_pickle('../results/'+name+'/'+base_dir+'/test_results.pkl')
| 0.351422 | 0.609669 |
# Ercot Summary Page - Calculations (LZ_SOUTH)
## Load in Data
```
import xlrd
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib
plt.style.use('ggplot')
# Set file path to Ercot Data
file_path = r"/Users/YoungFreeesh/Visual Studio Code/_Python/Web Scraping/Ercot/MASTER-Ercot.xlsx"
# read all data from "Master Data" tab from "MASTER-Ercot"
dfMASTER = pd.read_excel(file_path, sheet_name = 'Master Data')
# Convert df to a Date Frame
dfMASTER = pd.DataFrame(dfMASTER)
# Get Headers of "Master Data"
headers = list(dfMASTER.columns.values)
# Get Unique Months by creating an Array of the active worksheet names
xls = xlrd.open_workbook(file_path, on_demand=True)
SheetNameArray = xls.sheet_names()
UniqueMonths = SheetNameArray[3:]
### Refine the DataFrame
#Only Take LZ_SOUTH
dfMASTER_LZ_SOUTH = dfMASTER[['Oper Day', 'Interval Ending', 'LZ_SOUTH']].copy(deep=True)
dfMASTER_LZ_SOUTH['Oper Day'] = pd.to_datetime(dfMASTER_LZ_SOUTH['Oper Day'])
# Reset index to Oper Day
dfMASTER_LZ_SOUTH = dfMASTER_LZ_SOUTH.set_index('Oper Day')
dfMASTER_LZ_SOUTH.head(8)
```
## Begin Calculations
### 1) Avg. Daily Price
```
### Initialize dataframe
daily_avg = pd.DataFrame()
# Resample df & compute mean
daily_avg['Mean'] = dfMASTER_LZ_SOUTH.LZ_SOUTH.resample('D').mean()
daily_avg
```
### 2) Avg. Price - Power was cut off from 3:15pm to 4:15pm
Remove Prices from: 1515 to 1615 --> Remove (1530, 1545, 1600, 1615)
(i.e. remove those 4 data points and take the avg again)
```
# 1530 - 1615
dfMASTER_LZ_SOUTH_Optimized = dfMASTER_LZ_SOUTH[ ( (dfMASTER_LZ_SOUTH['Interval Ending'] <= 1515) | (dfMASTER_LZ_SOUTH['Interval Ending'] >= 1630) ) ].copy(deep=True)
# 1530 - 1715
dfMASTER_LZ_SOUTH_Optimized2 = dfMASTER_LZ_SOUTH[ ( (dfMASTER_LZ_SOUTH['Interval Ending'] <= 1515) | (dfMASTER_LZ_SOUTH['Interval Ending'] >= 1730) ) ].copy(deep=True)
#daily_avg_Optimized
### Initialize dataframe
daily_avg_Optimized = pd.DataFrame()
# Resample df & compute mean
daily_avg_Optimized['Mean'] = dfMASTER_LZ_SOUTH_Optimized.LZ_SOUTH.resample('D').mean()
#daily_avg_Optimized
daily_avg_Optimized2 = pd.DataFrame()
# Resample df & compute mean
daily_avg_Optimized2['Mean'] = dfMASTER_LZ_SOUTH_Optimized2.LZ_SOUTH.resample('D').mean()
#daily_avg_Optimized
```
### 3)Difference between (1) and (2)
```
difference = daily_avg - daily_avg_Optimized
difference2 = daily_avg - daily_avg_Optimized2
print(daily_avg.describe())
print(daily_avg_Optimized.describe())
print(daily_avg_Optimized2.describe())
print(difference.describe())
print(difference2.describe())
#difference
```
## Create Summary DataFrame
```
dfSummary = pd.DataFrame()
dfSummary['Avg Daily Price'] = daily_avg['Mean']
dfSummary['Avg Daily Price - Optimized'] = daily_avg_Optimized['Mean']
dfSummary['Avg Daily Price - Optimized 2'] = daily_avg_Optimized2['Mean']
dfSummary['Difference'] = difference['Mean']
dfSummary['Difference 2'] = difference2['Mean']
print('Avg Daily Price SUM: ', daily_avg['Mean'].sum())
print('Avg Daily Price - Optimized SUM: ', daily_avg_Optimized['Mean'].sum())
print('Avg Daily Price - Optimized 2 SUM: ', daily_avg_Optimized2['Mean'].sum())
print('Difference SUM: ', difference['Mean'].sum())
print('Difference 2 SUM: ', difference2['Mean'].sum())
dfSummary
```
|
github_jupyter
|
import xlrd
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib
plt.style.use('ggplot')
# Set file path to Ercot Data
file_path = r"/Users/YoungFreeesh/Visual Studio Code/_Python/Web Scraping/Ercot/MASTER-Ercot.xlsx"
# read all data from "Master Data" tab from "MASTER-Ercot"
dfMASTER = pd.read_excel(file_path, sheet_name = 'Master Data')
# Convert df to a Date Frame
dfMASTER = pd.DataFrame(dfMASTER)
# Get Headers of "Master Data"
headers = list(dfMASTER.columns.values)
# Get Unique Months by creating an Array of the active worksheet names
xls = xlrd.open_workbook(file_path, on_demand=True)
SheetNameArray = xls.sheet_names()
UniqueMonths = SheetNameArray[3:]
### Refine the DataFrame
#Only Take LZ_SOUTH
dfMASTER_LZ_SOUTH = dfMASTER[['Oper Day', 'Interval Ending', 'LZ_SOUTH']].copy(deep=True)
dfMASTER_LZ_SOUTH['Oper Day'] = pd.to_datetime(dfMASTER_LZ_SOUTH['Oper Day'])
# Reset index to Oper Day
dfMASTER_LZ_SOUTH = dfMASTER_LZ_SOUTH.set_index('Oper Day')
dfMASTER_LZ_SOUTH.head(8)
### Initialize dataframe
daily_avg = pd.DataFrame()
# Resample df & compute mean
daily_avg['Mean'] = dfMASTER_LZ_SOUTH.LZ_SOUTH.resample('D').mean()
daily_avg
# 1530 - 1615
dfMASTER_LZ_SOUTH_Optimized = dfMASTER_LZ_SOUTH[ ( (dfMASTER_LZ_SOUTH['Interval Ending'] <= 1515) | (dfMASTER_LZ_SOUTH['Interval Ending'] >= 1630) ) ].copy(deep=True)
# 1530 - 1715
dfMASTER_LZ_SOUTH_Optimized2 = dfMASTER_LZ_SOUTH[ ( (dfMASTER_LZ_SOUTH['Interval Ending'] <= 1515) | (dfMASTER_LZ_SOUTH['Interval Ending'] >= 1730) ) ].copy(deep=True)
#daily_avg_Optimized
### Initialize dataframe
daily_avg_Optimized = pd.DataFrame()
# Resample df & compute mean
daily_avg_Optimized['Mean'] = dfMASTER_LZ_SOUTH_Optimized.LZ_SOUTH.resample('D').mean()
#daily_avg_Optimized
daily_avg_Optimized2 = pd.DataFrame()
# Resample df & compute mean
daily_avg_Optimized2['Mean'] = dfMASTER_LZ_SOUTH_Optimized2.LZ_SOUTH.resample('D').mean()
#daily_avg_Optimized
difference = daily_avg - daily_avg_Optimized
difference2 = daily_avg - daily_avg_Optimized2
print(daily_avg.describe())
print(daily_avg_Optimized.describe())
print(daily_avg_Optimized2.describe())
print(difference.describe())
print(difference2.describe())
#difference
dfSummary = pd.DataFrame()
dfSummary['Avg Daily Price'] = daily_avg['Mean']
dfSummary['Avg Daily Price - Optimized'] = daily_avg_Optimized['Mean']
dfSummary['Avg Daily Price - Optimized 2'] = daily_avg_Optimized2['Mean']
dfSummary['Difference'] = difference['Mean']
dfSummary['Difference 2'] = difference2['Mean']
print('Avg Daily Price SUM: ', daily_avg['Mean'].sum())
print('Avg Daily Price - Optimized SUM: ', daily_avg_Optimized['Mean'].sum())
print('Avg Daily Price - Optimized 2 SUM: ', daily_avg_Optimized2['Mean'].sum())
print('Difference SUM: ', difference['Mean'].sum())
print('Difference 2 SUM: ', difference2['Mean'].sum())
dfSummary
| 0.485356 | 0.758287 |
# Importing from the Standard Library
## math module
The documentation for the `math` module is [here](https://docs.python.org/3/library/math.html)
Importing a single function from a module
```
from math import sqrt
answer = sqrt(2)
print(answer)
```
Importing the entire module
```
import math
pi = math.pi # modules can include constants (unchanging variable values)
print(pi)
answer = math.cos(pi)
print(answer)
```
Abbreviating an imported module
```
import math as m
answer = m.log10(1000)
print(answer)
# The same function can be referenced multiple ways
# depending on how it was previously imported
print(sqrt(2))
print(math.sqrt(2))
print(m.sqrt(2))
```
## time module
[documentation](https://docs.python.org/3/library/time.html)
```
# Import the time module
import time
# Print current local time as a formatted string
# "Local time" may be ambiguous if running in the cloud!
print(time.strftime('%H:%M:%S'))
print("I'm going to go to sleep for 3 seconds!")
# Suspend execution for 3 seconds
time.sleep(3)
print("I'm awake!")
print(time.strftime('%H:%M:%S'))
```
## os module
[documetation](https://docs.python.org/3/library/os.html)
```
import os
working_directory = os.getcwd()
print(working_directory)
print(os.listdir()) # no argument gets working directory
print(os.listdir(working_directory + '/Documents'))
```
# Methods
## String methods
[documentation](https://docs.python.org/3/library/string.html)
These methods operate on the built-in string class `str`.
`.upper()`, `.lower()`, and `.title()` methods have no arguments. They return a string.
```
my_message = 'Do not yell at me, Steve!'
shouting = my_message.upper()
print(shouting)
ee_cummings = my_message.lower()
print(ee_cummings)
my_book = my_message.title()
print(my_book)
```
## datetime module
The `datetime` module is part of the Standard Library.
[documentation](https://docs.python.org/3/library/datetime.html)
We introduce two new kinds of objects: date and datetime.
```
import datetime
# Instantiate two date objects, numeric arguments required.
sep_11 = datetime.date(2001,9,11)
this_day = datetime.date.today() # method sets the date value as today
print(type(sep_11))
print(sep_11.isoformat()) # use ISO 8601 format
print(sep_11.weekday()) # numeric value; Monday is 0
print(sep_11.strftime('%A')) # '%A' is a string format code for the day
print()
print(this_day.isoformat())
print(this_day.weekday())
print(this_day.strftime('%A'))
# Instantiate a dateTime object
# The dateTime will be expressed as Universal Coordinated Time (UTC)
# a.k.a. Greenwich Mean Time (GMT)
right_now = datetime.datetime.utcnow()
print(type(right_now))
print(right_now.isoformat())
# See the datetime module documentation for the string format codes
print(right_now.strftime('%B %d, %Y %I:%M %p'))
```
# Practice
```
# Import the time.sleep() function to use without a prefix
# Make the script sleep for 1 second
# Import the datetime module abbreviated as 'dt'
# Instantiate now as a date and dateTime using the abbreviation.
```
|
github_jupyter
|
from math import sqrt
answer = sqrt(2)
print(answer)
import math
pi = math.pi # modules can include constants (unchanging variable values)
print(pi)
answer = math.cos(pi)
print(answer)
import math as m
answer = m.log10(1000)
print(answer)
# The same function can be referenced multiple ways
# depending on how it was previously imported
print(sqrt(2))
print(math.sqrt(2))
print(m.sqrt(2))
# Import the time module
import time
# Print current local time as a formatted string
# "Local time" may be ambiguous if running in the cloud!
print(time.strftime('%H:%M:%S'))
print("I'm going to go to sleep for 3 seconds!")
# Suspend execution for 3 seconds
time.sleep(3)
print("I'm awake!")
print(time.strftime('%H:%M:%S'))
import os
working_directory = os.getcwd()
print(working_directory)
print(os.listdir()) # no argument gets working directory
print(os.listdir(working_directory + '/Documents'))
my_message = 'Do not yell at me, Steve!'
shouting = my_message.upper()
print(shouting)
ee_cummings = my_message.lower()
print(ee_cummings)
my_book = my_message.title()
print(my_book)
import datetime
# Instantiate two date objects, numeric arguments required.
sep_11 = datetime.date(2001,9,11)
this_day = datetime.date.today() # method sets the date value as today
print(type(sep_11))
print(sep_11.isoformat()) # use ISO 8601 format
print(sep_11.weekday()) # numeric value; Monday is 0
print(sep_11.strftime('%A')) # '%A' is a string format code for the day
print()
print(this_day.isoformat())
print(this_day.weekday())
print(this_day.strftime('%A'))
# Instantiate a dateTime object
# The dateTime will be expressed as Universal Coordinated Time (UTC)
# a.k.a. Greenwich Mean Time (GMT)
right_now = datetime.datetime.utcnow()
print(type(right_now))
print(right_now.isoformat())
# See the datetime module documentation for the string format codes
print(right_now.strftime('%B %d, %Y %I:%M %p'))
# Import the time.sleep() function to use without a prefix
# Make the script sleep for 1 second
# Import the datetime module abbreviated as 'dt'
# Instantiate now as a date and dateTime using the abbreviation.
| 0.341802 | 0.890865 |
<a href="https://colab.research.google.com/github/kapilkn/ML/blob/master/MNIST_Hand_Written_Kapil.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import tensorflow as tf
import matplotlib.pyplot as plt
import numpy as np
mnist = tf.keras.datasets.mnist
(xtrain,ytrain),(xtest,ytest)=mnist.load_data()
xtrain.shape,ytrain.shape
xtest.shape,ytest.shape
xtrain[10]
ytrain[10]
for i in range(25):
plt.subplot(5,5,i+1)
plt.imshow(xtrain[i])
plt.show()
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.imshow(xtrain[i])
plt.show()
plt.figure(figsize=(8,8))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xlabel(ytrain[i])
plt.xticks([])
plt.yticks([])
plt.imshow(xtrain[i],cmap='gray')
plt.show()
```
## Normaliztion
```
xtrain = tf.keras.utils.normalize(xtrain)
xtest = tf.keras.utils.normalize(xtest)
```
## Build the model
```
model = tf.keras.models.Sequential()
```
# Add layers
```
model.add(tf.keras.layers.Flatten()) # input layer (I dont know the dimension)
model.add(tf.keras.layers.Dense(784,activation='relu')) # hidden layer (28x28)
model.add(tf.keras.layers.Dense(600,activation='relu')) # hidden layer
model.add(tf.keras.layers.Dense(64,activation='relu')) # hidden layer
model.add(tf.keras.layers.Dense(10,activation='softmax')) # output layer
```
## Configure the model
```
model.compile(loss='sparse_categorical_crossentropy', optimizer='adam',metrics=['accuracy'])
model.fit(xtrain,ytrain,epochs=3)
predictions = model.predict(xtest)
tf.keras.utils.plot_model(
model, to_file='model.png', show_shapes=True, show_layer_names=True,
rankdir='LR', expand_nested=True
)
plt.figure(figsize=(8,8))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xlabel(np.argmax(predictions[i]))
plt.xticks([])
plt.yticks([])
plt.imshow(xtest[i],cmap='gray')
plt.show()
```
## Evaluate ANN
```
loss,accu = model.evaluate(xtest,ytest)
print(loss,accu)
from keras.models import Sequential
from keras.layers import Dense
from keras.utils.vis_utils import plot_model
plot_model(model, to_file='model_plot.png', show_shapes=True, show_layer_names=True)
from IPython.display import display, Javascript
from google.colab.output import eval_js
from base64 import b64decode
def take_photo(filename='photo.jpg', quality=0.8):
js = Javascript('''
async function takePhoto(quality) {
const div = document.createElement('div');
const capture = document.createElement('button');
capture.textContent = 'Take a Picture';
div.appendChild(capture);
const video = document.createElement('video');
video.style.display = 'block';
const stream = await navigator.mediaDevices.getUserMedia({video: true});
document.body.appendChild(div);
div.appendChild(video);
video.srcObject = stream;
await video.play();
// Resize the output to fit the video element.
google.colab.output.setIframeHeight(document.documentElement.scrollHeight, true);
// Wait for Capture to be clicked.
await new Promise((resolve) => capture.onclick = resolve);
const canvas = document.createElement('canvas');
canvas.width = video.videoWidth;
canvas.height = video.videoHeight;
canvas.getContext('2d').drawImage(video, 0, 0);
stream.getVideoTracks()[0].stop();
div.remove();
return canvas.toDataURL('image/jpeg', quality);
}
''')
display(js)
data = eval_js('takePhoto({})'.format(quality))
binary = b64decode(data.split(',')[1])
with open(filename, 'wb') as f:
f.write(binary)
return filename
from IPython.display import Image
try:
filename = take_photo()
print('Saved to {}'.format(filename))
# Show the image which was just taken.
display(Image(filename))
except Exception as err:
# Errors will be thrown if the user does not have a webcam or if they do not
# grant the page permission to access it.
print(str(err))
# Live image
from PIL import Image
import cv2
user_test = filename
col = Image.open(user_test)
gray = col.convert('L')
bw = gray.point(lambda x: 0 if x<100 else 255, '1')
bw.save("a.png")
bw
img_array = cv2.imread("a.png", cv2.IMREAD_GRAYSCALE)
img_array = cv2.bitwise_not(img_array)
print(img_array.size)
plt.imshow(img_array, cmap = plt.cm.binary)
plt.show()
img_size = 28
new_array = cv2.resize(img_array, (img_size,img_size))
plt.imshow(new_array, cmap = plt.cm.binary)
plt.show()
user_test = tf.keras.utils.normalize(new_array, axis = 1)
predicted = model.predict(np.array([[user_test]]))
a = predicted[0][0]
for i in range(0,10):
b = predicted[0][i]
print("Probability Distribution for",i,b)
print("The Predicted Value is",np.argmax(predicted[0]))
```
|
github_jupyter
|
import tensorflow as tf
import matplotlib.pyplot as plt
import numpy as np
mnist = tf.keras.datasets.mnist
(xtrain,ytrain),(xtest,ytest)=mnist.load_data()
xtrain.shape,ytrain.shape
xtest.shape,ytest.shape
xtrain[10]
ytrain[10]
for i in range(25):
plt.subplot(5,5,i+1)
plt.imshow(xtrain[i])
plt.show()
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.imshow(xtrain[i])
plt.show()
plt.figure(figsize=(8,8))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xlabel(ytrain[i])
plt.xticks([])
plt.yticks([])
plt.imshow(xtrain[i],cmap='gray')
plt.show()
xtrain = tf.keras.utils.normalize(xtrain)
xtest = tf.keras.utils.normalize(xtest)
model = tf.keras.models.Sequential()
model.add(tf.keras.layers.Flatten()) # input layer (I dont know the dimension)
model.add(tf.keras.layers.Dense(784,activation='relu')) # hidden layer (28x28)
model.add(tf.keras.layers.Dense(600,activation='relu')) # hidden layer
model.add(tf.keras.layers.Dense(64,activation='relu')) # hidden layer
model.add(tf.keras.layers.Dense(10,activation='softmax')) # output layer
model.compile(loss='sparse_categorical_crossentropy', optimizer='adam',metrics=['accuracy'])
model.fit(xtrain,ytrain,epochs=3)
predictions = model.predict(xtest)
tf.keras.utils.plot_model(
model, to_file='model.png', show_shapes=True, show_layer_names=True,
rankdir='LR', expand_nested=True
)
plt.figure(figsize=(8,8))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xlabel(np.argmax(predictions[i]))
plt.xticks([])
plt.yticks([])
plt.imshow(xtest[i],cmap='gray')
plt.show()
loss,accu = model.evaluate(xtest,ytest)
print(loss,accu)
from keras.models import Sequential
from keras.layers import Dense
from keras.utils.vis_utils import plot_model
plot_model(model, to_file='model_plot.png', show_shapes=True, show_layer_names=True)
from IPython.display import display, Javascript
from google.colab.output import eval_js
from base64 import b64decode
def take_photo(filename='photo.jpg', quality=0.8):
js = Javascript('''
async function takePhoto(quality) {
const div = document.createElement('div');
const capture = document.createElement('button');
capture.textContent = 'Take a Picture';
div.appendChild(capture);
const video = document.createElement('video');
video.style.display = 'block';
const stream = await navigator.mediaDevices.getUserMedia({video: true});
document.body.appendChild(div);
div.appendChild(video);
video.srcObject = stream;
await video.play();
// Resize the output to fit the video element.
google.colab.output.setIframeHeight(document.documentElement.scrollHeight, true);
// Wait for Capture to be clicked.
await new Promise((resolve) => capture.onclick = resolve);
const canvas = document.createElement('canvas');
canvas.width = video.videoWidth;
canvas.height = video.videoHeight;
canvas.getContext('2d').drawImage(video, 0, 0);
stream.getVideoTracks()[0].stop();
div.remove();
return canvas.toDataURL('image/jpeg', quality);
}
''')
display(js)
data = eval_js('takePhoto({})'.format(quality))
binary = b64decode(data.split(',')[1])
with open(filename, 'wb') as f:
f.write(binary)
return filename
from IPython.display import Image
try:
filename = take_photo()
print('Saved to {}'.format(filename))
# Show the image which was just taken.
display(Image(filename))
except Exception as err:
# Errors will be thrown if the user does not have a webcam or if they do not
# grant the page permission to access it.
print(str(err))
# Live image
from PIL import Image
import cv2
user_test = filename
col = Image.open(user_test)
gray = col.convert('L')
bw = gray.point(lambda x: 0 if x<100 else 255, '1')
bw.save("a.png")
bw
img_array = cv2.imread("a.png", cv2.IMREAD_GRAYSCALE)
img_array = cv2.bitwise_not(img_array)
print(img_array.size)
plt.imshow(img_array, cmap = plt.cm.binary)
plt.show()
img_size = 28
new_array = cv2.resize(img_array, (img_size,img_size))
plt.imshow(new_array, cmap = plt.cm.binary)
plt.show()
user_test = tf.keras.utils.normalize(new_array, axis = 1)
predicted = model.predict(np.array([[user_test]]))
a = predicted[0][0]
for i in range(0,10):
b = predicted[0][i]
print("Probability Distribution for",i,b)
print("The Predicted Value is",np.argmax(predicted[0]))
| 0.690663 | 0.930521 |
<center>
<img src="https://gitlab.com/ibm/skills-network/courses/placeholder101/-/raw/master/labs/module%201/images/IDSNlogo.png" width="300" alt="cognitiveclass.ai logo" />
</center>
# **SpaceX Falcon 9 First Stage Landing Prediction**
## Assignment: Exploring and Preparing Data
Estimated time needed: **70** minutes
In this assignment, we will predict if the Falcon 9 first stage will land successfully. SpaceX advertises Falcon 9 rocket launches on its website with a cost of 62 million dollars; other providers cost upward of 165 million dollars each, much of the savings is due to the fact that SpaceX can reuse the first stage.
In this lab, you will perform Exploratory Data Analysis and Feature Engineering.
Falcon 9 first stage will land successfully

Several examples of an unsuccessful landing are shown here:

Most unsuccessful landings are planned. Space X performs a controlled landing in the oceans.
## Objectives
Perform exploratory Data Analysis and Feature Engineering using `Pandas` and `Matplotlib`
* Exploratory Data Analysis
* Preparing Data Feature Engineering
***
### Import Libraries and Define Auxiliary Functions
We will import the following libraries the lab
```
# andas is a software library written for the Python programming language for data manipulation and analysis.
import pandas as pd
#NumPy is a library for the Python programming language, adding support for large, multi-dimensional arrays and matrices, along with a large collection of high-level mathematical functions to operate on these arrays
import numpy as np
# Matplotlib is a plotting library for python and pyplot gives us a MatLab like plotting framework. We will use this in our plotter function to plot data.
import matplotlib.pyplot as plt
#Seaborn is a Python data visualization library based on matplotlib. It provides a high-level interface for drawing attractive and informative statistical graphics
import seaborn as sns
```
## Exploratory Data Analysis
First, let's read the SpaceX dataset into a Pandas dataframe and print its summary
```
df=pd.read_csv("https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBM-DS0321EN-SkillsNetwork/datasets/dataset_part_2.csv")
# If you were unable to complete the previous lab correctly you can uncomment and load this csv
# df = pd.read_csv('https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-DS0701EN-SkillsNetwork/api/dataset_part_2.csv')
df.head(5)
```
First, let's try to see how the `FlightNumber` (indicating the continuous launch attempts.) and `Payload` variables would affect the launch outcome.
We can plot out the <code>FlightNumber</code> vs. <code>PayloadMass</code>and overlay the outcome of the launch. We see that as the flight number increases, the first stage is more likely to land successfully. The payload mass is also important; it seems the more massive the payload, the less likely the first stage will return.
```
sns.catplot(y="PayloadMass", x="FlightNumber", hue="Class", data=df, aspect = 5)
plt.xlabel("Flight Number",fontsize=20)
plt.ylabel("Pay load Mass (kg)",fontsize=20)
plt.show()
```
We see that different launch sites have different success rates. <code>CCAFS LC-40</code>, has a success rate of 60 %, while <code>KSC LC-39A</code> and <code>VAFB SLC 4E</code> has a success rate of 77%.
Next, let's drill down to each site visualize its detailed launch records.
### TASK 1: Visualize the relationship between Flight Number and Launch Site
Use the function <code>catplot</code> to plot <code>FlightNumber</code> vs <code>LaunchSite</code>, set the parameter <code>x</code> parameter to <code>FlightNumber</code>,set the <code>y</code> to <code>Launch Site</code> and set the parameter <code>hue</code> to <code>'class'</code>
```
# Plot a scatter point chart with x axis to be Flight Number and y axis to be the launch site, and hue to be the class value
sns.catplot(y="LaunchSite", x="FlightNumber", hue="Class", data=df, aspect = 5)
plt.xlabel("Flight Number",fontsize=20)
plt.ylabel("LaunchSite",fontsize=20)
plt.show()
```
Now try to explain the patterns you found in the Flight Number vs. Launch Site scatter point plots.
### TASK 2: Visualize the relationship between Payload and Launch Site
We also want to observe if there is any relationship between launch sites and their payload mass.
```
# Plot a scatter point chart with x axis to be Pay Load Mass (kg) and y axis to be the launch site, and hue to be the class value
sns.catplot(y="LaunchSite", x="PayloadMass", hue="Class", data=df, aspect = 5)
plt.xlabel("PayloadMass",fontsize=20)
plt.ylabel("LaunchSite",fontsize=20)
plt.show()
```
Conclusion: Large payloads appear to come from 2 sites only CCAFS and KSC.
### TASK 3: Visualize the relationship between success rate of each orbit type
Next, we want to visually check if there are any relationship between success rate and orbit type.
Let's create a `bar chart` for the sucess rate of each orbit
```
# HINT use groupby method on Orbit column and get the mean of Class column
orb_grp=df.groupby(['Orbit']).Class.mean()
# Plot bar chart
orb_grp.plot(kind="bar")
orb_grp
orb_grp=orb_grp.reset_index()
orb_grp
sns.barplot(orb_grp["Orbit"], orb_grp["Class"])
```
Analyze the ploted bar chart try to find which orbits have high sucess rate.
### TASK 4: Visualize the relationship between FlightNumber and Orbit type
For each orbit, we want to see if there is any relationship between FlightNumber and Orbit type.
```
# Plot a scatter point chart with x axis to be FlightNumber and y axis to be the Orbit, and hue to be the class value
sns.scatterplot(data=df, x='FlightNumber', y='Orbit', hue='Class', alpha=.5)
```
You should see that in the LEO orbit the Success appears related to the number of flights; on the other hand, there seems to be no relationship between flight number when in GTO orbit.
### TASK 5: Visualize the relationship between Payload and Orbit type
Similarly, we can plot the Payload vs. Orbit scatter point charts to reveal the relationship between Payload and Orbit type
```
# Plot a scatter point chart with x axis to be Payload and y axis to be the Orbit, and hue to be the class value
sns.scatterplot(data=df, x='PayloadMass', y='Orbit', hue='Class', alpha=.5)
```
You should observe that Heavy payloads have a negative influence on GTO orbits and positive on GTO and Polar LEO (ISS) orbits.
### TASK 6: Visualize the launch success yearly trend
You can plot a line chart with x axis to be <code>Year</code> and y axis to be average success rate, to get the average launch success trend.
The function will help you get the year from the date:
```
# A function to Extract years from the date
year=[]
def Extract_year(date):
for i in df["Date"]:
year.append(i.split("-")[0])
return year
year=Extract_year(df["Date"])
df['Year']=year
yr_grp=df.groupby(['Year']).Class.mean()
yr_grp=yr_grp.reset_index()
yr_grp
# Plot a line chart with x axis to be the extracted year and y axis to be the success rate
sns.lineplot(data=yr_grp, x="Year", y="Class")
df['LaunchSite'].value_counts()
df['Orbit'].value_counts()
df['Outcome'].value_counts()
```
you can observe that the sucess rate since 2013 kept increasing till 2020
## Features Engineering
By now, you should obtain some preliminary insights about how each important variable would affect the success rate, we will select the features that will be used in success prediction in the future module.
```
features = df[['FlightNumber', 'PayloadMass', 'Orbit', 'LaunchSite', 'Flights', 'GridFins', 'Reused', 'Legs', 'LandingPad', 'Block', 'ReusedCount', 'Serial']]
features.head()
```
### TASK 7: Create dummy variables to categorical columns
Use the function <code>get_dummies</code> and <code>features</code> dataframe to apply OneHotEncoder to the column <code>Orbits</code>, <code>LaunchSite</code>, <code>LandingPad</code>, and <code>Serial</code>. Assign the value to the variable <code>features_one_hot</code>, display the results using the method head. Your result dataframe must include all features including the encoded ones.
```
# HINT: Use get_dummies() function on the categorical columns
catg=df[df.select_dtypes(include=['object']).columns.tolist()]
catg2=catg[[ "Orbit", "LaunchSite", "LandingPad","Serial"]]
features_one_hot=pd.get_dummies(catg2)
features_one_hot
```
### TASK 8: Cast all numeric columns to `float64`
Now that our <code>features_one_hot</code> dataframe only contains numbers cast the entire dataframe to variable type <code>float64</code>
```
# HINT: use astype function
df[df.select_dtypes(exclude=['object','bool']).columns.tolist()].astype(float)
```
We can now export it to a <b>CSV</b> for the next section,but to make the answers consistent, in the next lab we will provide data in a pre-selected date range.
<code>features_one_hot.to_csv('dataset_part\_3.csv', index=False)</code>
## Authors
<a href="https://www.linkedin.com/in/joseph-s-50398b136/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDS0321ENSkillsNetwork26802033-2021-01-01">Joseph Santarcangelo</a> has a PhD in Electrical Engineering, his research focused on using machine learning, signal processing, and computer vision to determine how videos impact human cognition. Joseph has been working for IBM since he completed his PhD.
<a href="https://www.linkedin.com/in/nayefaboutayoun/?utm_medium=Exinfluencer&utm_source=Exinfluencer&utm_content=000026UJ&utm_term=10006555&utm_id=NA-SkillsNetwork-Channel-SkillsNetworkCoursesIBMDS0321ENSkillsNetwork26802033-2021-01-01">Nayef Abou Tayoun</a> is a Data Scientist at IBM and pursuing a Master of Management in Artificial intelligence degree at Queen's University.
## Change Log
| Date (YYYY-MM-DD) | Version | Changed By | Change Description |
| ----------------- | ------- | ---------- | ----------------------- |
| 2020-09-20 | 1.0 | Joseph | Modified Multiple Areas |
| 2020-11-10 | 1.1 | Nayef | updating the input data |
Copyright © 2020 IBM Corporation. All rights reserved.
|
github_jupyter
|
# andas is a software library written for the Python programming language for data manipulation and analysis.
import pandas as pd
#NumPy is a library for the Python programming language, adding support for large, multi-dimensional arrays and matrices, along with a large collection of high-level mathematical functions to operate on these arrays
import numpy as np
# Matplotlib is a plotting library for python and pyplot gives us a MatLab like plotting framework. We will use this in our plotter function to plot data.
import matplotlib.pyplot as plt
#Seaborn is a Python data visualization library based on matplotlib. It provides a high-level interface for drawing attractive and informative statistical graphics
import seaborn as sns
df=pd.read_csv("https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBM-DS0321EN-SkillsNetwork/datasets/dataset_part_2.csv")
# If you were unable to complete the previous lab correctly you can uncomment and load this csv
# df = pd.read_csv('https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-DS0701EN-SkillsNetwork/api/dataset_part_2.csv')
df.head(5)
sns.catplot(y="PayloadMass", x="FlightNumber", hue="Class", data=df, aspect = 5)
plt.xlabel("Flight Number",fontsize=20)
plt.ylabel("Pay load Mass (kg)",fontsize=20)
plt.show()
# Plot a scatter point chart with x axis to be Flight Number and y axis to be the launch site, and hue to be the class value
sns.catplot(y="LaunchSite", x="FlightNumber", hue="Class", data=df, aspect = 5)
plt.xlabel("Flight Number",fontsize=20)
plt.ylabel("LaunchSite",fontsize=20)
plt.show()
# Plot a scatter point chart with x axis to be Pay Load Mass (kg) and y axis to be the launch site, and hue to be the class value
sns.catplot(y="LaunchSite", x="PayloadMass", hue="Class", data=df, aspect = 5)
plt.xlabel("PayloadMass",fontsize=20)
plt.ylabel("LaunchSite",fontsize=20)
plt.show()
# HINT use groupby method on Orbit column and get the mean of Class column
orb_grp=df.groupby(['Orbit']).Class.mean()
# Plot bar chart
orb_grp.plot(kind="bar")
orb_grp
orb_grp=orb_grp.reset_index()
orb_grp
sns.barplot(orb_grp["Orbit"], orb_grp["Class"])
# Plot a scatter point chart with x axis to be FlightNumber and y axis to be the Orbit, and hue to be the class value
sns.scatterplot(data=df, x='FlightNumber', y='Orbit', hue='Class', alpha=.5)
# Plot a scatter point chart with x axis to be Payload and y axis to be the Orbit, and hue to be the class value
sns.scatterplot(data=df, x='PayloadMass', y='Orbit', hue='Class', alpha=.5)
# A function to Extract years from the date
year=[]
def Extract_year(date):
for i in df["Date"]:
year.append(i.split("-")[0])
return year
year=Extract_year(df["Date"])
df['Year']=year
yr_grp=df.groupby(['Year']).Class.mean()
yr_grp=yr_grp.reset_index()
yr_grp
# Plot a line chart with x axis to be the extracted year and y axis to be the success rate
sns.lineplot(data=yr_grp, x="Year", y="Class")
df['LaunchSite'].value_counts()
df['Orbit'].value_counts()
df['Outcome'].value_counts()
features = df[['FlightNumber', 'PayloadMass', 'Orbit', 'LaunchSite', 'Flights', 'GridFins', 'Reused', 'Legs', 'LandingPad', 'Block', 'ReusedCount', 'Serial']]
features.head()
# HINT: Use get_dummies() function on the categorical columns
catg=df[df.select_dtypes(include=['object']).columns.tolist()]
catg2=catg[[ "Orbit", "LaunchSite", "LandingPad","Serial"]]
features_one_hot=pd.get_dummies(catg2)
features_one_hot
# HINT: use astype function
df[df.select_dtypes(exclude=['object','bool']).columns.tolist()].astype(float)
| 0.70253 | 0.986044 |
# Single network model theory
## Foundation
To understand network models, it is crucial to understand the concept of a network as a random quantity, taking a probability distribution. We have a realization $A$, and we think that this realization is random in some way. Stated another way, we think that there exists a network-valued random variable $\mathbf A$ that governs the realizations we get to see. Since $\mathbf A$ is a random variable, we can describe it using a probability distribution. The distribution of the random network $\mathbf A$ is the function $\mathbb P$ which assigns probabilities to every possible configuration that $\mathbf A$ could take. Notationally, we write that $\mathbf A \sim \mathbb P$, which is read in words as "the random network $\mathbf A$ is distributed according to $\mathbb P$."
In the preceding description, we made a fairly substantial claim: $\mathbb P$ assigns probabilities to every possible configuration that realizations of $\mathbf A$, denoted by $A$, could take. How many possibilities are there for a network with $n$ nodes? Let's limit ourselves to simple networks: that is, $A$ takes values that are unweighted ($A$ is *binary*), undirected ($A$ is *symmetric*), and loopless ($A$ is *hollow*). In words, $\mathcal A_n$ is the set of all possible adjacency matrices $A$ that correspond to simple networks with $n$ nodes. Stated another way: every $A$ that is found in $\mathcal A$ is a *binary* $n \times n$ matrix ($A \in \{0, 1\}^{n \times n}$), $A$ is symmetric ($A = A^\top$), and $A$ is *hollow* ($diag(A) = 0$, or $A_{ii} = 0$ for all $i = 1,...,n$). We describe $\mathcal A_n$ as:
\begin{align*}
\mathcal A_n = \left\{A : A \textrm{ is an $n \times n$ matrix with $0$s and $1$s}, A\textrm{ is symmetric}, A\textrm{ is hollow}\right\}
\end{align*}
To summarize the statement that $\mathbb P$ assigns probabilities to every possible configuration that realizations of $\mathbf A$ can take, we write that $\mathbb P : \mathcal A_n \rightarrow [0, 1]$. This means that for any $A \in \mathcal A_n$ which is a possible realization of a random network $\mathbf A$, that $\mathbb P(\mathbf A = A)$ is a probability (it takes a value between $0$ and $1$). If it is completely unambiguous what the random variable $\mathbf A$ refers to, we might abbreviate $\mathbb P(\mathbf A = A)$ with $\mathbb P(A)$. This statement can alternatively be read that the probability that the random variable $\mathbf A$ takes the value $A$ is $\mathbb P(A)$. Finally, let's address that question we had in the previous paragraph. How many possible adjacency matrices are in $\mathcal A_n$?
Let's imagine what just one $A \in \mathcal A_n$ can look like. Note that each matrix $A$ has $n \times n = n^2$ possible entries, in total, since $A$ is an $n \times n$ matrix. There are $n$ possible self-loops for a network, but since $\mathbf A$ is simple, it is loopless. This means that we can subtract $n$ possible edges from $n^2$, leaving us with $n^2 - n = n(n-1)$ possible edges that might not be unconnected. If we think in terms of a realization $A$, this means that we are ignoring the diagonal entries $a_{ii}$, for all $i \in [n]$. Remember that a simple network is also undirected. In terms of the realization $A$, this means that for every pair $i$ and $j$, that $a_{ij} = a_{ji}$. If we were to learn about an entry in the upper triangle of $A$ where $a_{ij}$ is such that $j > i$, note that we have also learned what $a_{ji}$ is, too. This symmetry of $A$ means that of the $n(n-1)$ entries that are not on the diagonal of $A$, we would, in fact, "double count" the possible number of unique values that $A$ could have. This means that $A$ has a total of $\frac{1}{2}n(n - 1)$ possible entries which are *free*, which is equal to the expression $\binom{n}{2}$. Finally, note that for each entry of $A$, that the adjacency can take one of two possible values: $0$ or $1$. To write this down formally, for every possible edge which is randomly determined, we have *two* possible values that edge could take. Let's think about building some intuition here:
1. If $A$ is $2 \times 2$, there are $\binom{2}{2} = 1$ unique entry of $A$, which takes one of $2$ values. There are $2$ possible ways that $A$ could look:
\begin{align*}
\begin{bmatrix}
0 & 1 \\
1 & 0
\end{bmatrix}\textrm{ or }
\begin{bmatrix}
0 & 0 \\
0 & 0
\end{bmatrix}
\end{align*}
2. If $A$ is $3 \times 3$, there are $\binom{3}{2} = \frac{3 \times 2}{2} = 3$ unique entries of $A$, each of which takes one of $2$ values. There are $8$ possible ways that $A$ could look:
\begin{align*}
&\begin{bmatrix}
0 & 1 & 1 \\
1 & 0 & 1 \\
1 & 1 & 0
\end{bmatrix}\textrm{ or }
\begin{bmatrix}
0 & 1 & 0 \\
1 & 0 & 1 \\
0 & 1 & 0
\end{bmatrix}\textrm{ or }
\begin{bmatrix}
0 & 0 & 1 \\
0 & 0 & 1 \\
1 & 1 & 0
\end{bmatrix}
\textrm{ or }\\
&\begin{bmatrix}
0 & 1 & 1 \\
1 & 0 & 0 \\
1 & 0 & 0
\end{bmatrix}\textrm{ or }
\begin{bmatrix}
0 & 0 & 1 \\
0 & 0 & 0 \\
1 & 0 & 0
\end{bmatrix}\textrm{ or }
\begin{bmatrix}
0 & 0 & 0 \\
0 & 0 & 1 \\
0 & 1 & 0
\end{bmatrix}\textrm{ or }\\
&\begin{bmatrix}
0 & 1 & 0 \\
1 & 0 & 0 \\
0 & 0 & 0
\end{bmatrix}\textrm{ or }
\begin{bmatrix}
0 & 0 & 0 \\
0 & 0 & 0 \\
0 & 0 & 0
\end{bmatrix}
\end{align*}
How do we generalize this to an arbitrary choice of $n$? The answer is to use *combinatorics*. Basically, the approach is to look at each entry of $A$ which can take different values, and multiply the total number of possibilities by $2$ for every element which can take different values. Stated another way, if there are $2$ choices for each one of $x$ possible items, we have $2^x$ possible ways in which we could select those $x$ items. But we already know how many different elements there are in $A$, so we are ready to come up with an expression for the number. In total, there are $2^{\binom n 2}$ unique adjacency matrices in $\mathcal A_n$. Stated another way, the *cardinality* of $\mathcal A_n$, described by the expression $|\mathcal A_n|$, is $2^{\binom n 2}$. The **cardinality** here just means the number of elements that the set $\mathcal A_n$ contains. When $n$ is just $15$, note that $\left|\mathcal A_{15}\right| = 2^{\binom{15}{2}} = 2^{105}$, which when expressed as a power of $10$, is more than $10^{30}$ possible networks that can be realized with just $15$ nodes! As $n$ increases, how many unique possible networks are there? In the below figure, look at the value of $|\mathcal A_n| = 2^{\binom n 2}$ as a function of $n$. As we can see, as $n$ gets big, $|\mathcal A_n|$ grows really really fast!
```
import seaborn as sns
import numpy as np
from math import comb
n = np.arange(2, 51)
logAn = np.array([comb(ni, 2) for ni in n])*np.log10(2)
ax = sns.lineplot(x=n, y=logAn)
ax.set_title("")
ax.set_xlabel("Number of Nodes")
ax.set_ylabel("Number of Possible Graphs $|A_n|$ (log scale)")
ax.set_yticks([50, 100, 150, 200, 250, 300, 350])
ax.set_yticklabels(["$10^{{{pow:d}}}$".format(pow=d) for d in [50, 100, 150, 200, 250, 300, 350]])
ax;
```
So, now we know that we have probability distributions on networks, and a set $\mathcal A_n$ which defines all of the adjacency matrices that every probability distribution must assign a probability to. Now, just what is a network model? A **network model** is a set $\mathcal P$ of probability distributions on $\mathcal A_n$. Stated another way, we can describe $\mathcal P$ to be:
\begin{align*}
\mathcal P &\subseteq \{\mathbb P: \mathbb P\textrm{ is a probability distribution on }\mathcal A_n\}
\end{align*}
In general, we will simplify $\mathcal P$ through something called *parametrization*. We define $\Theta$ to be the set of all possible parameters of the random network model, and $\theta \in \Theta$ is a particular parameter choice that governs the parameters of a specific network-valued random variaable $\mathbf A$. In this case, we will write $\mathcal P$ as the set:
\begin{align*}
\mathcal P(\Theta) &= \left\{\mathbb P_\theta : \theta \in \Theta\right\}
\end{align*}
If $\mathbf A$ is a random network that follows a network model, we will write that $\mathbf A \sim \mathbb P_\theta$, for some choice $\theta$. We will often use the shorthand $\mathbf A \sim \mathbb P$.
If you are used to traditional univariate or multivariate statistical modelling, an extremely natural choice for when you have a discrete sample space (like $\mathcal A_n$, which is discrete because we can count it) would be to use a categorical model. In the categorical model, we would have a single parameter for all possible configurations of an $n$-node network; that is, $|\theta| = \left|\mathcal A_n\right| = 2^{\binom n 2}$. What is wrong with this model? The limitations are two-fold:
1. As we explained previously, when $n$ is just $15$, we would need over $10^{30}$ bits of storage just to define $\theta$. This amounts to more than $10^{8}$ zetabytes, which exceeds the storage capacity of *the entire world*.
2. With a single network observed (or really, any number of networks we could collect in the real world) we would never be able to get a reasonable estimate of $2^{\binom n 2}$ parameters for any reasonably non-trivial number of nodes $n$. For the case of one observed network $A$, an estimate of $\theta$ (referred to as $\hat\theta$) would simply be for $\hat\theta$ to have a $1$ in the entry corresponding to our observed network, and a $0$ everywhere else. Inferentially, this would imply that the network-valued random variable $\mathbf A$ which governs realizations $A$ is deterministic, even if this is not the case. Even if we collected potentially *many* observed networks, we would still (with very high probability) just get $\hat \theta$ as a series of point masses on the observed networks we see, and $0$s everywhere else. This would mean our parameter estimates $\hat\theta$ would not generalize to new observations at *all*, with high probability.
So, what are some more reasonable descriptions of $\mathcal P$? We explore some choices below. Particularly, we will be most interested in the *independent-edge* networks. These are the families of networks in which the generative procedure which governs the random networks assume that the edges of the network are generated *independently*. **Statistical Independence** is a property which greatly simplifies many of the modelling assumptions which are crucial for proper estimation and rigorous statistical inference, which we will learn more about in the later chapters.
### Equivalence Classes
In all of the below models, we will explore the concept of the **probability equivalence class**, or an *equivalence class*, for short. The probability is a function which in general, describes how effective a particular observation can be described by a random variable $\mathbf A$ with parameters $\theta$, written $\mathbf A \sim F(\theta)$. The probability will be used to describe the probability $\mathbb P_\theta(\mathbf A)$ of observing the realization $A$ if the underlying random variable $\mathbf A$ has parameters $\theta$. Why does this matter when it comes to equivalence classes? An equivalence class is a subset of the sample space $E \subseteq \mathcal A_n$, which has the following properties. Holding the parameters $\theta$ fixed:
1. If $A$ and $A'$ are members of the same equivalence class $E$ (written $A, A' \in E$), then $\mathbb P_\theta(A) = \mathbb P_\theta(A')$.
2. If $A$ and $A''$ are members of different equivalence classes; that is, $A \in E$ and $A'' \in E'$ where $E, E'$ are equivalence classes, then $\mathbb P_\theta(A) \neq \mathbb P_\theta(A'')$.
3. Using points 1 and 2, we can establish that if $E$ and $E'$ are two different equivalence classes, then $E \cap E' = \varnothing$. That is, the equivalence classes are **mutually disjoint**.
4. We can use the preceding properties to deduce that given the sample space $\mathcal A_n$ and a probability function $\mathbb P_\theta$, we can define a partition of the sample space into equivalence classes $E_i$, where $i \in \mathcal I$ is an arbitrary indexing set. A **partition** of $\mathcal A_n$ is a sequence of sets which are mutually disjoint, and whose union is the whole space. That is, $\bigcup_{i \in \mathcal I} E_i = \mathcal A_n$.
We will see more below about how the equivalence classes come into play with network models, and in a later section, we will see their relevance to the estimation of the parameters $\theta$.
(representations:whyuse:networkmodels:iern)=
### Independent-Edge Random Networks
The below models are all special families of something called **independent-edge random networks**. An independent-edge random network is a network-valued random variable, in which the collection of edges are all independent. In words, this means that for every adjacency $\mathbf a_{ij}$ of the network-valued random variable $\mathbf A$, that $\mathbf a_{ij}$ is independent of $\mathbf a_{i'j'}$, any time that $(i,j) \neq (i',j')$. When the networks are simple, the easiest thing to do is to assume that each edge $(i,j)$ is connected with some probability (which might be different for each edge) $p_{ij}$. We use the $ij$ subscript to denote that this probability is not necessarily the same for each edge. This simple model can be described as $\mathbf a_{ij}$ has the distribution $Bern(p_{ij})$, for every $j > i$, and is independent of every other edge in $\mathbf A$. We only look at the entries $j > i$, since our networks are simple. This means that knowing a realization of $\mathbf a_{ij}$ also gives us the realizaaion of $\mathbf a_{ji}$ (and thus $\mathbf a_{ji}$ is a *deterministic* function of $\mathbf a_{ij}$). Further, we know that the random network is loopless, which means that every $\mathbf a_{ii} = 0$. We will call the matrix $P = (p_{ij})$ the **probability matrix** of the network-valued random variable $\mathbf A$. In general, we will see a common theme for the probabilities of a realization $A$ of a network-valued random variable $\mathbf A$, which is that it will greatly simplify our computation. Remember that if $\mathbf x$ and $\mathbf y$ are binary variables which are independent, that $\mathbb P(\mathbf x = x, \mathbf y = y) = \mathbb P(\mathbf x = x) \mathbb P(\mathbf y = y)$. Using this fact:
\begin{align*}
\mathbb P(\mathbf A = A) &= \mathbb P(\mathbf a_{11} = a_{11}, \mathbf a_{12} = a_{12}, ..., \mathbf a_{nn} = a_{nn}) \\
&= \mathbb P(\mathbf a_{ij} = a_{ij} \text{ for all }j > i) \\
&= \prod_{j > i}\mathbb P(\mathbf a_{ij} = a_{ij}), \;\;\;\;\textrm{Independence Assumption}
\end{align*}
Next, we will use the fact that if a random variable $\mathbf a_{ij}$ has the Bernoulli distribution with probability $p_{ij}$, that $\mathbb P(\mathbf a_{ij} = a_{ij}) = p_{ij}^{a_{ij}}(1 - p_{ij})^{1 - p_{ij}}$:
\begin{align*}
\mathbb P_\theta(A) &= \prod_{j > i}p_{ij}^{a_{ij}}(1 - p_{ij})^{1 - p_{ij}}
\end{align*}
Now that we've specified a probability and a very generalizable model, we've learned the full story behind network models and are ready to skip to estimating parameters, right? *Wrong!* Unfortunately, if we tried too estimate anything about each $p_{ij}$ individually, we would obtain that $p_{ij} = a_{ij}$ if we only have one realization $A$. Even if we had many realizations of $\mathbf A$, this still would not be very interesting, since we have a *lot* of $p_{ij}$s to estimate, and we've ignored any sort of structural model that might give us deeper insight into $\mathbf A$. In the below sections, we will learn successively less restrictive (and hence, *more expressive*) assumptions about $p_{ij}$s, which will allow us to convey fairly complex random networks, but *still* enable us with plenty of intteresting things to learn about later on.
## Erdös-Rényi (ER) Random Networks
The Erdös Rényi model formalizes this relatively simple situation with a single parameter and an $iid$ assumption:
| Parameter | Space | Description |
| --- | --- | --- |
| $p$ | $[0, 1]$ | Probability that an edge exists between a pair of nodes, which is identical for all pairs of nodes |
From here on out, when we talk about an Erdös Rényi random variable, we will simply call it an ER network. In an ER network, each pair of nodes is connected with probability $p$, and therefore not connected with probability $1-p$. Statistically, we say that for each edge $\mathbf{a}_{ij}$ for every pair of nodes where $j > i$ (in terms of the adjacency matrix, this means all of the edges in the *upper right* triangle), that $\mathbf{a}_{ij}$ is sampled independently and identically from a *Bernoulli* distribution with probability $p$. The word "independent" means that edges in the network occurring or not occurring do not affect one another. For instance, this means that if we knew a student named Alice was friends with Bob, and Alice was also friends with Chadwick, that we do not learn any information about whether Bob is friends with Chadwick. The word "identical" means that every edge in the network has the same probability $p$ of being connected. If Alice and Bob are friends with probability $p$, then Alice and Chadwick are friends with probability $p$, too. We assume here that the networks are undirected, which means that if an edge $\mathbf a_{ij}$ exists from node $i$ to $j$, then the edge $\mathbf a_{ji}$ also exists from node $j$ to node $i$. We also assume that the networks are loopless, which means that no edges $\mathbf a_{ii}$ can go from node $i$ to itself. If $\mathbf A$ is the adjacency matrix for an ER network with probability $p$, we write that $\mathbf A \sim ER_n(p)$.
Next, let's formalize an example of one of the limitations of an ER random network. Remember that we said that ER random networks are often too simple. Well, one way in which they are simple is called **degree homogeneity**, which is a property in which *all* of the nodes in an ER network have the *exact* same expected node degree! What this means is that if we were to take an ER random network $\mathbf A$, we would expect that *all* of the nodes in the network had the same degree. Let's see how this works:
```{admonition} Working Out the Expected Degree in an Erdös-Rényi Network
Suppose that $\mathbf A$ is a simple network which is random. The network has $n$ nodes $\mathcal V = (v_i)_{i = 1}^n$. Recall that the in a simple network, the node degree is $deg(v_i) = \sum_{j = 1}^n \mathbf a_{ij}$. What is the expected degree of a node $v_i$ of a random network $\mathbf A$ which is Erdös-Rényi?
To describe this, we will compute the expectated value of the degree $deg(v_i)$, written $\mathbb E\left[deg(v_i)\right]$. Let's see what happens:
\begin{align*}
\mathbb E\left[deg(v_i)\right] &= \mathbb E\left[\sum_{j = 1}^n \mathbf a_{ij}\right] \\
&= \sum_{j = 1}^n \mathbb E[\mathbf a_{ij}]
\end{align*}
We use the *linearity of expectation* in the line above, which means that the expectation of a sum with a finite number of terms being summed over ($n$, in this case) is the sum of the expectations. Finally, by definition, all of the edges $A_{ij}$ have the same distribution: $Bern(p)$. The expected value of a random quantity which takes a Bernoulli distribution is just the probability $p$. This means every term $\mathbb E[\mathbf a_{ij}] = p$. Therefore:
\begin{align*}
\mathbb E\left[deg(v_i)\right] &= \sum_{j = 1}^n p = n\cdot p
\end{align*}
Since all of the $n$ terms being summed have the same expected value. This holds for *every* node $v_i$, which means that the expected degree of all nodes is an undirected ER network is the same number, $n \cdot p$.
```
### Probability
What is the probability for realizations of Erdös-Rényi networks? Remember that for Independent-edge graphs, that the probability can be written:
\begin{align*}
\mathbb P_{\theta}(A) &= \prod_{j > i} \mathbb P_\theta(\mathbf{a}_{ij} = a_{ij})
\end{align*}
Next, we recall that by assumption of the ER model, that the probability matrix $P = (p)$, or that $p_{ij} = p$ for all $i,j$. Therefore:
\begin{align*}
\mathbb P_\theta(A) &= \prod_{j > i} p^{a_{ij}}(1 - p)^{1 - a_{ij}} \\
&= p^{\sum_{j > i} a_{ij}} \cdot (1 - p)^{\binom{n}{2} - \sum_{j > i}a_{ij}} \\
&= p^{m} \cdot (1 - p)^{\binom{n}{2} - m}
\end{align*}
This means that the probability $\mathbb P_\theta(A)$ is a function *only* of the number of edges $m = \sum_{j > i}a_{ij}$ in the network represented by adjacency matrix $A$. The equivalence class on the Erdös-Rényi networks are the sets:
\begin{align*}
E_{i} &= \left\{A \in \mathcal A_n : m = i\right\}
\end{align*}
where $i$ index from $0$ (the minimum number of edges possible) all the way up to $n^2$ (the maximum number of edges possible). All of the relationships for equivalence classes discussed above apply to the sets $E_i$.
## Network Models for networks which aren't simple
To make the discussions a little more easy to handle, in the above descriptions and all our successive descriptions, we will describe network models for **simple networks**. To recap, networks which are simple are binary networks which are both loopless and undirected. Stated another way, simple networks are networks whose adjacency matrices are only $0$s and $1$s, they are hollow (the diagonal is entirely *0*), and symmetric (the lower and right triangles of the adjacency matrix are the *same*). What happens our networks don't quite look this way?
For now, we'll keep the assumption that the networks are binary, but we will discuss non-binary network models in a later chapter. We have three possibilities we can consider, and we will show how the "relaxations" of the assumptions change a description of a network model. A *relaxation*, in statistician speak, means that we are taking the assumptions that we had (in this case, that the networks are *simple*), and progressively making the assumptions weaker (more *relaxed*) so that they apply to other networks, too. We split these out so we can be as clear as possible about how the generative model changes with each relaxation step.
We will compare each relaxation to the statement about the generative model for the ER generative model. To recap, for a simple network, we wrote:
"Statistically, we say that for each edge $\mathbf{a}_{ij}$ for every pair of nodes where $j > i$ (in terms of the adjacency matrix, this means all of the nodes in the *upper right* triangle), that $\mathbf{a}_{ij}$ is sampled independently and identically from a *Bernoulli* distribution with probability $p$.... We assume here that the networks are undirected, which means that if an edge $\mathbf a_{ij}$ exists from node $i$ to $j$, then the edge $\mathbf a_{ji}$ also exists from node $j$ to node $i$. We also assume that the networks are loopless, which means that no edges $\mathbf a_{ii}$ can go from node $i$ to itself."
Any additional parts that are added are expressed in **<font color='green'>green</font>** font. Omitted parts are struck through with <font color='red'><strike>red</strike></font> font.
Note that these generalizations apply to *any* of the successive networks which we describe in the Network Models section, and not just the ER model!
### Binary network model which has loops, but is undirected
Here, all we want to do is relax the assumption that the network is loopless. We simply ignore the statement that edges $\mathbf a_{ii}$ cannot exist, and allow that the $\mathbf a_{ij}$ which follow a Bernoulli distribution (with some probability which depends on the network model choice) *now* applies to $j \geq i$, and not just $j > i$. We keep that an edge $\mathbf a_{ij}$ existing implies that $\mathbf a_{ji}$ also exists, which maintains the symmetry of $\mathbf A$ (and consequently, the undirectedness of the network).
Our description of the ER network changes to:
Statistically, we say that for each edge $\mathbf{a}_{ij}$ for every pair of nodes where $\mathbf{\color{green}{j \geq i}}$ (in terms of the adjacency matrix, this means all of the nodes in the *upper right* triangle **<font color='green'>and the diagonal</font>**), that $\mathbf{a}_{ij}$ is sampled independently and identically from a *Bernoulli* distribution with probability $p$.... We assume here that the networks are undirected, which means that if an edge $\mathbf a_{ij}$ exists from node $i$ to $j$, then the edge $\mathbf a_{ji}$ also exists from node $j$ to node $i$. <font color='red'><strike>We also assume that the networks are loopless, which means that no edges $\mathbf a_{ii}$ can go from node $i$ to itself.</strike></font>
### Binary network model which is loopless, but directed
Like above, we simply ignore the statement that $\mathbf a_{ji} = \mathbf a_{ij}$, which removes the symmetry of $\mathbf A$ (and consequently, removes the undirectedness of the network). We allow that the $\mathbf a_{ij}$ which follows a Bernoulli distribution now apply to $j \neq i$, and not just $j > i$. We keep that $\mathbf a_{ii} = 0$, which maintains the hollowness of $\mathbf A$ (and consequently, the undirectedness of the network).
Our description of the ER network changes to:
Statistically, we say that for each edge $\mathbf{a}_{ij}$ for every pair of nodes where $\mathbf{\color{green}{j \neq i}}$ (in terms of the adjacency matrix, this means all of the nodes <strike><font color='red'>in the *upper right* triangle</font></strike>**<font color='green'>which are not along the diagonal</font>**), that $\mathbf{a}_{ij}$ is sampled independently and identically from a *Bernoulli* distribution with probability $p$.... <font color='red'><strike>We assume here that the networks are undirected, which means that if an edge $\mathbf a_{ij}$ exists from node $i$ to $j$, then the edge $\mathbf a_{ji}$ also exists from node $j$ to node $i$.</strike></font> We also assume that the networks are loopless, which means that no edges $\mathbf a_{ii}$ can go from node $i$ to itself.
### Binary network model which is has loops and is directed
Finally, for a network which has loops and is directed, we combine the above two approaches. We ignore the statements that $\mathbf a_{ji} = \mathbf a_{ij}$, and the statement that $\mathbf a_{ii} = 0$.
Our descriptiomn of the ER network changes to:
Statistically, we say that for each edge $\mathbf{a}_{ij}$ <font color='red'><strike>where $j > i$ (in terms of the adjacency matrix, this means all of the nodes in the *upper right* triangle)</strike></font>, that $\mathbf{a}_{ij}$ is sampled independently and identically from a *Bernoulli* distribution with probability $p$, <font color='green'>for all possible combinations of nodes $j$ and $i$</font>. <font color='red'><strike>We assume here that the networks are undirected, which means that if an edge $\mathbf a_{ij}$ exists from node $i$ to $j$, then the edge $\mathbf a_{ji}$ also exists from node $j$ to node $i$. We also assume that the networks are loopless, which means that no edges $\mathbf a_{ii}$ can go from node $i$ to itself.</strike></font>
## *A Priori* Stochastic Block Model
The *a priori* SBM is an SBM in which we know ahead of time (*a priori*) which nodes are in which communities. Here, we will use the variable $K$ to denote the maximum number of different communities. The ordering of the communities does not matter; the community we call $1$ versus $2$ versus $K$ is largely a symbolic distinction (the only thing that matters is that they are *different*). The *a priori* SBM has the following parameter:
| Parameter | Space | Description |
| --- | --- | --- |
| $B$ | [0,1]$^{K \times K}$ | The block matrix, which assigns edge probabilities for pairs of communities |
To describe the *A Priori* SBM, we will designate the community each node is a part of using a vector, which has a single community assignment for each node in the network. We will call this **node assignment vector** $\vec{\tau}$, and it is a $n$-length vector (one element for each node) with elements which can take values from $1$ to $K$. In symbols, we would say that $\vec\tau \in \{1, ..., K\}^n$. What this means is that for a given element of $\vec \tau$, $\tau_i$, that $\tau_i$ is the community assignment (either $1$, $2$, so on and so forth up to $K$) for the $i^{th}$ node. If there we hahd an example where there were $2$ communities ($K = 2$) for instance, and the first two nodes are in community $1$ and the second two in community $2$, then $\vec\tau$ would be a vector which looks like:
\begin{align*}
\vec\tau &= \begin{bmatrix}1 & 1 & 2 & 2\end{bmatrix}^\top
\end{align*}
Next, let's discuss the matrix $B$, which is known as the **block matrix** of the SBM. We write down that $B \in [0, 1]^{K \times K}$, which means that the block matrix is a matrix with $K$ rows and $K$ columns. If we have a pair of nodes and know which of the $K$ communities each node is from, the block matrix tells us the probability that those two nodes are connected. If our networks are simple, the matrix $B$ is also symmetric, which means that if $b_{kk'} = p$ where $p$ is a probability, that $b_{k'k} = p$, too. The requirement of $B$ to be symmetric exists *only* if we are dealing with undirected networks.
Finally, let's think about how to write down the generative model for the *a priori* SBM. Intuitionally what we want to reflect is, if we know that node $i$ is in community $k'$ and node $j$ is in community $k$, that the $(k', k)$ entry of the block matrix is the probability that $i$ and $j$ are connected. We say that given $\tau_i = k'$ and $\tau_j = k$, $\mathbf a_{ij}$ is sampled independently from a $Bern(b_{k' k})$ distribution for all $j > i$. Note that the adjacencies $\mathbf a_{ij}$ are not *necessarily* identically distributed, because the probability depends on the community of edge $(i,j)$. If $\mathbf A$ is an *a priori* SBM network with parameter $B$, and $\vec{\tau}$ is a realization of the node-assignment vector, we write that $\mathbf A \sim SBM_{n,\vec \tau}(B)$.
### Probability
What does the probability for the *a priori* SBM look like? In our previous description, we admittedly simplified things to an extent to keep the wording down. In truth, we model the *a priori* SBM using a *latent variable* model, which means that the node assignment vector, $\vec{\pmb \tau}$, is treated as *random*. For the case of the *a priori* SBM, it just so happens that we *know* the specific value that this latent variable $\vec{\pmb \tau}$ takes, $\vec \tau$, ahead of time.
Fortunately, since $\vec \tau$ is a *parameter* of the *a priori* SBM, the probability is a bit simpler than for the *a posteriori* SBM. This is because the *a posteriori* SBM requires an integration over potential realizations of $\vec{\pmb \tau}$, whereas the *a priori* SBM does not, since we already know that $\vec{\pmb \tau}$ was realized as $\vec\tau$.
Putting these steps together gives us that:
\begin{align*}
\mathbb P_\theta(A) &= \mathbb P_{\theta}(\mathbf A = A | \vec{\pmb \tau} = \vec\tau) \\
&= \prod_{j > i} \mathbb P_\theta(\mathbf a_{ij} = a_{ij} | \vec{\pmb \tau} = \vec\tau),\;\;\;\;\textrm{Independence Assumption}
\end{align*}
Next, for the *a priori* SBM, we know that each edge $\mathbf a_{ij}$ only *actually* depends on the community assignments of nodes $i$ and $j$, so we know that $\mathbb P_{\theta}(\mathbf a_{ij} = a_{ij} | \vec{\pmb \tau} = \vec\tau) = \mathbb P(\mathbf a_{ij} = a_{ij} | \tau_i = k', \tau_j = k)$, where $k$ and $k'$ are any of the $K$ possible communities. This is because the community assignments of nodes that are not nodes $i$ and $j$ do not matter for edge $ij$, due to the independence assumption.
Next, let's think about the probability matrix $P = (p_{ij})$ for the *a priori* SBM. We know that, given that $\tau_i = k'$ and $\tau_j = k$, each adjacency $\mathbf a_{ij}$ is sampled independently and identically from a $Bern(b_{k',k})$ distribution. This means that $p_{ij} = b_{k',k}$. Completing our analysis from above:
\begin{align*}
\mathbb P_\theta(A) &= \prod_{j > i} b_{k'k}^{a_{ij}}(1 - b_{k'k})^{1 - a_{ij}} \\
&= \prod_{k,k' \in [K]}b_{k'k}^{m_{k'k}}(1 - b_{k'k})^{n_{k'k} - m_{k'k}}
\end{align*}
Where $n_{k' k}$ denotes the total number of edges possible between nodes assigned to community $k'$ and nodes assigned to community $k$. That is, $n_{k' k} = \sum_{j > i} \mathbb 1_{\tau_i = k'}\mathbb 1_{\tau_j = k}$. Further, we will use $m_{k' k}$ to denote the total number of edges observed between these two communities. That is, $m_{k' k} = \sum_{j > i}\mathbb 1_{\tau_i = k'}\mathbb 1_{\tau_j = k}a_{ij}$. Note that for a single $(k',k)$ community pair, that the probability is analogous to the probability of a realization of an ER random variable.
<!--- We can formalize this a bit more explicitly. If we let $A^{\ell k}$ be defined as the subgraph *induced* by the edges incident nodes in community $\ell$ and those in community $k$, then we can say that $A^{\ell k}$ is a directed ER random network, --->
Like the ER model, there are again equivalence classes of the sample space $\mathcal A_n$ in terms of their probability. For a two-community setting, with $\vec \tau$ and $B$ given, the equivalence classes are the sets:
\begin{align*}
E_{a,b,c}(\vec \tau, B) &= \left\{A \in \mathcal A_n : m_{11} = a, m_{21}=m_{12} = b, m_{22} = c\right\}
\end{align*}
The number of equivalence classes possible scales with the number of communities, and the manner in which nodes are assigned to communities (particularly, the number of nodes in each community).
## *A Posteriori* Stochastic Block Model
In the *a posteriori* Stochastic Block Model (SBM), we consider that node assignment to one of $K$ communities is a random variable, that we *don't* know already like te *a priori* SBM. We're going to see a funky word come up, that you're probably not familiar with, the **$K$ probability simplex**. What the heck is a probability simplex?
The intuition for a simplex is probably something you're very familiar with, but just haven't seen a word describe. Let's say I have a vector, $\vec\pi = (\pi_k)_{k \in [K]}$, which has a total of $K$ elements. $\vec\pi$ will be a vector, which indicates the *probability* that a given node is assigned to each of our $K$ communities, so we need to impose some additional constraints. Symbolically, we would say that, for all $i$, and for all $k$:
\begin{align*}
\pi_k = \mathbb P(\pmb\tau_i = k)
\end{align*}
The $\vec \pi$ we're going to use has a very special property: all of its elements are non-negative: for all $\pi_k$, $\pi_k \geq 0$. This makes sense since $\pi_k$ is being used to represent the probability of a node $i$ being in group $k$, so it certainly can't be negative. Further, there's another thing that we want our $\vec\pi$ to have: in order for each element $\pi_k$ to indicate the probability of something to be assigned to $k$, we need all of the $\pi_k$s to sum up to one. This is because of something called the Law of Total Probability. If we have $K$ total values that $\pmb \tau_i$ could take, then it is the case that:
\begin{align*}
\sum_{k=1}^K \mathbb P(\pmb \tau_i = k) = \sum_{k = 1}^K \pi_k = 1
\end{align*}
So, back to our question: how does a probability simplex fit in? Well, the $K$ probability simplex describes all of the possible values that our vector $\vec\pi$ could take! In symbols, the $K$ probability simplex is:
\begin{align*}
\left\{\vec\pi : \text{for all $k$ }\pi_k \geq 0, \sum_{k = 1}^K \pi_k = 1 \right\}
\end{align*}
So the $K$ probability simplex is just the space for all possible vectors which could indicate assignment probabilities to one of $K$ communities.
What does the probability simplex look like? Below, we take a look at the $2$-probability simplex (2-d $\vec\pi$s) and the $3$-probability simplex (3-dimensional $\vec\pi$s):
```
from mpl_toolkits.mplot3d import Axes3D
from mpl_toolkits.mplot3d.art3d import Poly3DCollection
import matplotlib.pyplot as plt
fig=plt.figure(figsize=plt.figaspect(.5))
fig.suptitle("Probability Simplexes")
ax=fig.add_subplot(1,2,1)
x=[1,0]
y=[0,1]
ax.plot(x,y)
ax.set_xticks([0,.5,1])
ax.set_yticks([0,.5,1])
ax.set_xlabel("$\pi_1$")
ax.set_ylabel("$\pi_2$")
ax.set_title("2-probability simplex")
ax=fig.add_subplot(1,2,2,projection='3d')
x = [1,0,0]
y = [0,1,0]
z = [0,0,1]
verts = [list(zip(x,y,z))]
ax.add_collection3d(Poly3DCollection(verts, alpha=.6))
ax.view_init(elev=20,azim=10)
ax.set_xticks([0,.5,1])
ax.set_yticks([0,.5,1])
ax.set_zticks([0,.5,1])
ax.set_xlabel("$\pi_1$")
ax.set_ylabel("$\pi_2$")
h=ax.set_zlabel("$\pi_3$", rotation=0)
ax.set_title("3-probability simplex")
plt.show()
```
The values of $\vec\pi = (\pi)$ that are in the $K$-probability simplex are indicated by the shaded region of each figure. This comprises the $(\pi_1, \pi_2)$ pairs that fall along a diagonal line from $(0,1)$ to $(1,0)$ for the $2$-simplex, and the $(\pi_1, \pi_2, \pi_3)$ tuples that fall on the surface of the triangular shape above with nodes at $(1,0,0)$, $(0,1,0)$, and $(0,0,1)$.
This model has the following parameters:
| Parameter | Space | Description |
| --- | --- | --- |
| $\vec \pi$ | the $K$ probability simplex | The probability of a node being assigned to community $K$ |
| $B$ | [0,1]$^{K \times K}$ | The block matrix, which assigns edge probabilities for pairs of communities |
The *a posteriori* SBM is a bit more complicated than the *a priori* SBM. We will think about the *a posteriori* SBM as a variation of the *a priori* SBM, where instead of the node-assignment vector being treated as a known fixed value (the community assignments), we will treat it as *unknown*. $\vec{\pmb \tau}$ is called a *latent variable*, which means that it is a quantity that is never actually observed, but which will be useful for describing our model. In this case, $\vec{\pmb \tau}$ takes values in the space $\{1,...,K\}^n$. This means that for a given realization of $\vec{\pmb \tau}$, denoted by $\vec \tau$, that for each of the $n$ nodes in the network, we suppose that an integer value between $1$ and $K$ indicates which community a node is from. Statistically, we write that the node assignment for node $i$, denoted by $\pmb \tau_i$, is sampled independently and identically from $Categorical(\vec \pi)$. Stated another way, the vector $\vec\pi$ indicates the probability $\pi_k$ of assignment to each community $k$ in the network.
The matrix $B$ behaves exactly the same as it did with the *a posteriori* SBM. Finally, let's think about how to write down the generative model in the *a posteriori* SBM. The model for the *a posteriori* SBM is, in fact, nearly the same as for the *a priori* SBM: we still say that given $\tau_i = k'$ and $\tau_j = k$, that $\mathbf a_{ij}$ are independent $Bern(b_{k'k})$. Here, however, we also describe that $\pmb \tau_i$ are sampled independent and identically from $Categorical(\vec\pi)$, as we learned above. If $\mathbf A$ is the adjacency matrix for an *a posteriori* SBM network with parameters $\vec \pi$ and $B$, we write that $\mathbf A \sim SBM_n(\vec \pi, B)$.
### Probability
What does the probability for the *a posteriori* SBM look like? In this case, $\theta = (\vec \pi, B)$ are the parameters for the model, so the probability for a realization $A$ of $\mathbf A$ is:
\begin{align*}
\mathbb P_\theta(A) &= \mathbb P_\theta(\mathbf A = A)
\end{align*}
Next, we use the fact that the probability that $\mathbf A = A$ is, in fact, the *integration* (over realizations of $\vec{\pmb \tau}$) of the joint $(\mathbf A, \vec{\pmb \tau})$. In this case, we will let $\mathcal T = \{1,...,K\}^n$ be the space of all possible realizations that $\vec{\pmb \tau}$ could take:
\begin{align}
\mathbb P_\theta(A)&= \sum_{\vec \tau \in \mathcal T} \mathbb P_\theta(\mathbf A = A, \vec{\pmb \tau} = \vec \tau)
\end{align}
Next, remember that by definition of a conditional probability for a random variable $\mathbf x$ taking value $x$ conditioned on random variable $\mathbf y$ taking the value $y$, that $\mathbb P(\mathbf x = x | \mathbf y = y) = \frac{\mathbb P(\mathbf x = x, \mathbf y = y)}{\mathbb P(\mathbf y = y)}$. Note that by multiplying through by $\mathbf P(\mathbf y = y)$, we can see that $\mathbb P(\mathbf x = x, \mathbf y = y) = \mathbb P(\mathbf x = x| \mathbf y = y)\mathbb P(\mathbf y = y)$. Using this logic for $\mathbf A$ and $\vec{\pmb \tau}$:
\begin{align*}
\mathbb P_\theta(A) &=\sum_{\vec \tau \in \mathcal T} \mathbb P_\theta(\mathbf A = A| \vec{\pmb \tau} = \vec \tau)\mathbb P(\vec{\pmb \tau} = \vec \tau)
\end{align*}
Intuitively, for each term in the sum, we are treating $\vec{\pmb \tau}$ as taking a fixed value, $\vec\tau$, to evaluate this probability statement.
We will start by describing $\mathbb P(\vec{\pmb \tau} = \vec\tau)$. Remember that for $\vec{\pmb \tau}$, that each entry $\pmb \tau_i$ is sampled *independently and identically* from $Categorical(\vec \pi)$.The probability mass for a $Categorical(\vec \pi)$-valued random variable is $\mathbb P(\pmb \tau_i = \tau_i; \vec \pi) = \pi_{\tau_i}$. Finally, note that if we are taking the products of $n$ $\pi_{\tau_i}$ terms, that many of these values will end up being the same. Consider, for instance, if the vector $\tau = [1,2,1,2,1]$. We end up with three terms of $\pi_1$, and two terms of $\pi_2$, and it does not matter which order we multiply them in. Rather, all we need to keep track of are the counts of each $\pi$ term. Written another way, we can use the indicator that $\tau_i = k$, given by $\mathbb 1_{\tau_i = k}$, and a running counter over all of the community probability assignments $\pi_k$ to make this expression a little more sensible. We will use the symbol $n_k = \sum_{i = 1}^n \mathbb 1_{\tau_i = k}$ to denote this value, which is the number of nodes in community $k$:
\begin{align*}
\mathbb P_\theta(\vec{\pmb \tau} = \vec \tau) &= \prod_{i = 1}^n \mathbb P_\theta(\pmb \tau_i = \tau_i),\;\;\;\;\textrm{Independence Assumption} \\
&= \prod_{i = 1}^n \pi_{\tau_i} ,\;\;\;\;\textrm{p.m.f. of a Categorical R.V.}\\
&= \prod_{k = 1}^K \pi_{k}^{n_k},\;\;\;\;\textrm{Reorganizing what we are taking products of}
\end{align*}
Next, let's think about the conditional probability term, $\mathbb P_\theta(\mathbf A = A \big | \vec{\pmb \tau} = \vec \tau)$. Remember that the entries are all independent conditional on $\vec{\pmb \tau}$ taking the value $\vec\tau$. It turns out this is exactly the same result that we obtained for the *a priori* SBM:
\begin{align*}
\mathbb P_\theta(\mathbf A = A \big | \vec{\pmb \tau} = \vec \tau)
&= \prod_{k',k} b_{\ell k}^{m_{k' k}}(1 - b_{k' k})^{n_{k' k} - m_{k' k}}
\end{align*}
Combining these into the integrand gives:
\begin{align*}
\mathbb P_\theta(A) &= \sum_{\vec \tau \in \mathcal T} \mathbb P_\theta(\mathbf A = A \big | \vec{\pmb \tau} = \vec \tau) \mathbb P_\theta(\vec{\pmb \tau} = \vec \tau) \\
&= \sum_{\vec \tau \in \mathcal T} \prod_{k = 1}^K \left[\pi_k^{n_k}\cdot \prod_{k'=1}^K b_{k' k}^{m_{k' k}}(1 - b_{k' k})^{n_{k' k} - m_{k' k}}\right]
\end{align*}
Evaluating this sum explicitly proves to be relatively tedious and is a bit outside of the scope of this book, so we will omit it here.
## Degree-Corrected Stochastic Block Model (DCSBM)
Let's think back to our school example for the Stochastic Block Model. Remember, we had 100 students, each of whom could go to one of two possible schools: school one or school two. Our network had 100 nodes, representing each of the students. We said that the school for which each student attended was represented by their node assignment $\tau_i$ to one of two possible communities. The matrix $B$ was the block probaability matrix, where $b_{11}$ was the probability that students in school one were friends, $b_{22}$ was the probability that students in school two were friends, and $b_{12} = b_{21}$ was the probability that students were friends if they did not go to the same school. In this case, we said that $\mathbf A$ was an $SBM_n(\tau, B)$ random network.
When would this setup not make sense? Let's say that Alice and Bob both go to the same school, but Alice is more popular than Bob. In general since Alice is more popular than Bob, we might want to say that for any clasasmate, Alice gets an additional "popularity benefit" to her probability of being friends with the other classmate, and Bob gets an "unpopularity penalty." The problem here is that within a single community of an SBM, the SBM assumes that the **node degree** (the number of nodes each nodes is connected to) is the *same* for all nodes within a single community. This means that we would be unable to reflect this benefit/penalty system to Alice and Bob, since each student will have the same number of friends, on average. This problem is referred to as **community degree homogeneity** in a Stochastic Block Model Network. Community degree homogeneity just means that the node degree is *homogeneous*, or the same, for all nodes within a community.
```{admonition} Degree Homogeneity in a Stochastic Block Model Network
Suppose that $\mathbf A \sim SBM_{n, \vec\tau}(B)$, where $\mathbf A$ has $K=2$ communities. What is the node degree of each node in $\mathbf A$?
For an arbitrary node $v_i$ which is in community $k$ (either one or two), we will compute the expectated value of the degree $deg(v_i)$, written $\mathbb E\left[deg(v_i); \tau_i = k\right]$. We will let $n_k$ represent the number of nodes whose node assignments $\tau_i$ are to community $k$. Let's see what happens:
\begin{align*}
\mathbb E\left[deg(v_i); \tau_i = k\right] &= \mathbb E\left[\sum_{j = 1}^n \mathbf a_{ij}\right] \\
&= \sum_{j = 1}^n \mathbb E[\mathbf a_{ij}]
\end{align*}
We use the *linearity of expectation* again to get from the top line to the second line. Next, instead of summing over all the nodes, we'll break the sum up into the nodes which are in the same community as node $i$, and the ones in the *other* community $k'$. We use the notation $k'$ to emphasize that $k$ and $k'$ are different values:
\begin{align*}
\mathbb E\left[deg(v_i); \tau_i = k\right] &= \sum_{j : i \neq j, \tau_j = k} \mathbb E\left[\mathbf a_{ij}\right] + \sum_{j : \tau_j =k'} \mathbb E[\mathbf a_{ij}]
\end{align*}
In the first sum, we have $n_k-1$ total edges (the number of nodes that aren't node $i$, but are in the same community), and in the second sum, we have $n_{k'}$ total edges (the number of nodes that are in the other community). Finally, we will use that the probability of an edge in the same community is $b_{kk}$, but the probability of an edge between the communities is $b_{k' k}$. Finally, we will use that the expected value of an adjacency $\mathbf a_{ij}$ which is Bernoulli distributed is its probability:
\begin{align*}
\mathbb E\left[deg(v_i); \tau_i = k\right] &= \sum_{j : i \neq j, \tau_j = k} b_{kk} + \sum_{j : \tau_j = \ell} b_{kk'},\;\;\;\;\mathbf a_{ij}\textrm{ are Bernoulli distributed} \\
&= (n_k - 1)b_{kk} + n_{k'} b_{kk'}
\end{align*}
This holds for any node $i$ which is in community $k$. Therefore, the expected node degree is the same, or **homogeneous**, within a community of an SBM.
```
To address this limitation, we turn to the Degree-Corrected Stochastic Block Model, or DCSBM. As with the Stochastic Block Model, there is both a *a priori* and *a posteriori* DCSBM.
### *A Priori* DCSBM
Like the *a priori* SBM, the *a priori* DCSBM is where we know which nodes are in which communities ahead of time. Here, we will use the variable $K$ to denote the number of different communiies. The *a priori* DCSBM has the following two parameters:
| Parameter | Space | Description |
| --- | --- | --- |
| $B$ | [0,1]$^{K \times K}$ | The block matrix, which assigns edge probabilities for pairs of communities |
| $\vec\theta$ | $\mathbb R^n_+$ | The degree correction vector, which adjusts the degree for pairs of nodes |
The latent community assignment vector $\vec{\pmb \tau}$ with a known *a priori* realization $\vec{\tau}$ and the block matrix $B$ are exactly the same for the *a priori* DCSBM as they were for the *a priori* SBM.
The vector $\vec\theta$ is the degree correction vector. Each entry $\theta_i$ is a positive scalar. $\theta_i$ defines how much more (or less) edges associated with node $i$ are connected due to their association with node $i$.
Finally, let's think about how to write down the generative model for the *a priori* DCSBM. We say that $\tau_i = k'$ and $\tau_j = k$, $\mathbf a_{ij}$ is sampled independently from a $Bern(\theta_i \theta_j b_{k'k})$ distribution for all $j > i$. As we can see, $\theta_i$ in a sense is "correcting" the probabilities of each adjacency to node $i$ to be higher, or lower, depending on the value of $\theta_i$ that that which is given by the block probabilities $b_{\ell k}$. If $\mathbf A$ is an *a priori* DCSBM network with parameters and $B$, we write that $\mathbf A \sim DCSBM_{n,\vec\tau}(\vec \theta, B)$.
#### Probability
The derivation for the probability is the same as for the *a priori* SBM, with the change that $p_{ij} = \theta_i \theta_j b_{k'k}$ instead of just $b_{k'k}$. This gives that the probability turns out to be:
\begin{align*}
\mathbb P_\theta(A) &= \prod_{j > i} \left(\theta_i \theta_j b_{k'k}\right)^{a_{ij}}\left(1 - \theta_i \theta_j b_{k'k}\right)^{1 - a_{ij}}
\end{align*}
The expression doesn't simplify much more due to the fact that the probabilities are dependent on the particular $i$ and $j$, so we can't just reduce the statement in terms of $n_{k'k}$ and $m_{k'k}$ like for the SBM.
### *A Posteriori* DCSBM
The *a posteriori* DCSBM is to the *a posteriori* SBM what the *a priori* DCSBM was to the *a priori* SBM. The changes are very minimal, so we will omit explicitly writing it all down here so we can get this section wrapped up, with the idea that the preceding section on the *a priori* DCSBM should tell you what needs to change. We will leave it as an exercise to the reader to write down a model and probability statement for realizations of the DCSBM.
## Random Dot Product Graph (RDPG)
### *A Priori* RDPG
The *a priori* Random Dot Product Graph is an RDPG in which we know *a priori* the latent position matrix $X$. The *a priori* RDPG has the following parameter:
| Parameter | Space | Description |
| --- | --- | --- |
| $X$ | $ \mathbb R^{n \times d}$ | The matrix of latent positions for each node $n$. |
$X$ is called the **latent position matrix** of the RDPG. We write that $X \in \mathbb R^{n \times d}$, which means that it is a matrix with real values, $n$ rows, and $d$ columns. We will use the notation $\vec x_i$ to refer to the $i^{th}$ row of $X$. $\vec x_i$ is referred to as the **latent position** of a node $i$. This looks something like this:
\begin{align*}
X = \begin{bmatrix}
\vec x_{1}^\top \\
\vdots \\
\vec x_n^\top
\end{bmatrix}
\end{align*}
Noting that $X$ has $d$ columns, this implies that $\vec x_i \in \mathbb R^d$, or that each node's latent position is a real-valued $d$-dimensional vector.
What is the generative model for the *a priori* RDPG? As we discussed above, given $X$, for all $j > i$, $\mathbf a_{ij} \sim Bern(\vec x_i^\top \vec x_j)$ independently. If $i < j$, $\mathbf a_{ji} = \mathbf a_{ij}$ (the network is *undirected*), and $\mathbf a_{ii} = 0$ (the network is *loopless*). If $\mathbf A$ is an *a priori* RDPG with parameter $X$, we write that $\mathbf A \sim RDPG_n(X)$.
<!-- TODO: return to add equivalence classes -->
#### Probability
Given $X$, the probability for an RDPG is relatively straightforward, as an RDPG is another Independent-Edge Random Graph. The independence assumption vastly simplifies our resulting expression. We will also use many of the results we've identified above, such as the p.m.f. of a Bernoulli random variable. Finally, we'll note that the probability matrix $P = (\vec x_i^\top \vec x_j)$, so $p_{ij} = \vec x_i^\top \vec x_j$:
\begin{align*}
\mathbb P_\theta(A) &= \mathbb P_\theta(A) \\
&= \prod_{j > i}\mathbb P(\mathbf a_{ij} = a_{ij}),\;\;\;\; \textrm{Independence Assumption} \\
&= \prod_{j > i}(\vec x_i^\top \vec x_j)^{a_{ij}}(1 - \vec x_i^\top \vec x_j)^{1 - a_{ij}},\;\;\;\; a_{ij} \sim Bern(\vec x_i^\top \vec x_j)
\end{align*}
Unfortunately, the probability equivalence classes are a bit harder to understand intuitionally here compared to the ER and SBM examples so we won't write them down here, but they still exist!
### *A Posteriori* RDPG
Like for the *a posteriori* SBM, the *a posteriori* RDPG introduces another strange set: the **intersection of the unit ball and the non-negative orthant**. Huh? This sounds like a real mouthful, but it turns out to be rather straightforward. You are probably already very familiar with a particular orthant: in two-dimensions, an orthant is called a quadrant. Basically, an orthant just extends the concept of a quadrant to spaces which might have more than $2$ dimensions. The non-negative orthant happens to be the orthant where all of the entries are non-negative. We call the **$K$-dimensional non-negative orthant** the set of points in $K$-dimensional real space, where:
\begin{align*}
\left\{\vec x \in \mathbb R^K : x_k \geq 0\text{ for all $k$}\right\}
\end{align*}
In two dimensions, this is the traditional upper-right portion of the standard coordinate axis. To give you a picture, the $2$-dimensional non-negative orthant is the blue region of the following figure:
```
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.axisartist import SubplotZero
import matplotlib.patches as patch
class myAxes():
def __init__(self, xlim=(-5,5), ylim=(-5,5), figsize=(6,6)):
self.xlim = xlim
self.ylim = ylim
self.figsize = figsize
self.__scale_arrows()
def __drawArrow(self, x, y, dx, dy, width, length):
plt.arrow(
x, y, dx, dy,
color = 'k',
clip_on = False,
head_width = self.head_width,
head_length = self.head_length
)
def __scale_arrows(self):
""" Make the arrows look good regardless of the axis limits """
xrange = self.xlim[1] - self.xlim[0]
yrange = self.ylim[1] - self.ylim[0]
self.head_width = min(xrange/30, 0.25)
self.head_length = min(yrange/30, 0.3)
def __drawAxis(self):
"""
Draws the 2D cartesian axis
"""
# A subplot with two additional axis, "xzero" and "yzero"
# corresponding to the cartesian axis
ax = SubplotZero(self.fig, 1, 1, 1)
self.fig.add_subplot(ax)
# make xzero axis (horizontal axis line through y=0) visible.
for axis in ["xzero","yzero"]:
ax.axis[axis].set_visible(True)
# make the other axis (left, bottom, top, right) invisible
for n in ["left", "right", "bottom", "top"]:
ax.axis[n].set_visible(False)
# Plot limits
plt.xlim(self.xlim)
plt.ylim(self.ylim)
ax.set_yticks([-1, 1, ])
ax.set_xticks([-2, -1, 0, 1, 2])
# Draw the arrows
self.__drawArrow(self.xlim[1], 0, 0.01, 0, 0.3, 0.2) # x-axis arrow
self.__drawArrow(0, self.ylim[1], 0, 0.01, 0.2, 0.3) # y-axis arrow
self.ax=ax
def draw(self):
# First draw the axis
self.fig = plt.figure(figsize=self.figsize)
self.__drawAxis()
axes = myAxes(xlim=(-2.5,2.5), ylim=(-2,2), figsize=(9,7))
axes.draw()
rectangle =patch.Rectangle((0,0), 3, 3, fc='blue',ec="blue", alpha=.2)
axes.ax.add_patch(rectangle)
plt.show()
```
Now, what is the unit ball? You are probably familiar with the idea of the unit ball, even if you haven't heard it called that specifically. Remember that the Euclidean norm for a point $\vec x$ which has coordinates $x_i$ for $i=1,...,K$ is given by the expression:
\begin{align*}
\left|\left|\vec x\right|\right|_2 = \sqrt{\sum_{i = 1}^K x_i^2}
\end{align*}
The Euclidean unit ball is just the set of points whose Euclidean norm is at most $1$. To be more specific, the **closed unit ball** with the Euclidean norm is the set of points:
\begin{align*}
\left\{\vec x \in \mathbb R^K :\left|\left|\vec x\right|\right|_2 \leq 1\right\}
\end{align*}
We draw the $2$-dimensional unit ball with the Euclidean norm below, where the points that make up the unit ball are shown in red:
```
axes = myAxes(xlim=(-2.5,2.5), ylim=(-2,2), figsize=(9,7))
axes.draw()
circle =patch.Circle((0,0), 1, fc='red',ec="red", alpha=.3)
axes.ax.add_patch(circle)
plt.show()
```
Now what is their intersection? Remember that the intersection of two sets $A$ and $B$ is the set:
\begin{align*}
A \cap B &= \{x : x \in A, x \in B\}
\end{align*}
That is, each element must be in *both* sets to be in the intersection. The interesction of the unit ball and the non-negative orthant will be the set:
\begin{align*}
\mathcal X_K = \left\{\vec x \in \mathbb R^K :\left|\left|\vec x\right|\right|_2 \leq 1, x_k \geq 0 \textrm{ for all $k$}\right\}
\end{align*}
visually, this will be the set of points in the *overlap* of the unit ball and the non-negative orthant, which we show below in purple:
```
axes = myAxes(xlim=(-2.5,2.5), ylim=(-2,2), figsize=(9,7))
axes.draw()
circle =patch.Circle((0,0), 1, fc='red',ec="red", alpha=.3)
axes.ax.add_patch(circle)
rectangle =patch.Rectangle((0,0), 3, 3, fc='blue',ec="blue", alpha=.2)
axes.ax.add_patch(rectangle)
plt.show()
```
This space has an *incredibly* important corollary. It turns out that if $\vec x$ and $\vec y$ are both elements of $\mathcal X_K$, that $\left\langle \vec x, \vec y \right \rangle = \vec x^\top \vec y$, the **inner product**, is at most $1$, and at least $0$. Without getting too technical, this is because of something called the Cauchy-Schwartz inequality and the properties of $\mathcal X_K$. If you remember from linear algebra, the Cauchy-Schwartz inequality states that $\left\langle \vec x, \vec y \right \rangle$ can be at most the product of $\left|\left|\vec x\right|\right|_2$ and $\left|\left|\vec y\right|\right|_2$. Since $\vec x$ and $\vec y$ have norms both less than or equal to $1$ (since they are on the *unit ball*), their inner-product is at most $1$. Further, since $\vec x$ and $\vec y$ are in the non-negative orthant, their inner product can never be negative. This is because both $\vec x$ and $\vec y$ have entries which are not negative, and therefore their element-wise products can never be negative.
The *a posteriori* RDPG is to the *a priori* RDPG what the *a posteriori* SBM was to the *a priori* SBM. We instead suppose that we do *not* know the latent position matrix $X$, but instead know how we can characterize the individual latent positions. We have the following parameter:
| Parameter | Space | Description |
| --- | --- | --- |
| F | inner-product distributions | A distribution which governs each latent position. |
The parameter $F$ is what is known as an **inner-product distribution**. In the simplest case, we will assume that $F$ is a distribution on a subset of the possible real vectors that have $d$-dimensions with an important caveat: for any two vectors within this subset, their inner product *must* be a probability. We will refer to the subset of the possible real vectors as $\mathcal X_K$, which we learned about above. This means that for any $\vec x_i, \vec x_j$ that are in $\mathcal X_K$, it is always the case that $\vec x_i^\top \vec x_j$ is between $0$ and $1$. This is essential because like previously, we will describe the distribution of each edge in the adjacency matrix using $\vec x_i^\top \vec x_j$ to represent a probability. Next, we will treat the latent position matrix as a matrix-valued random variable which is *latent* (remember, *latent* means that we don't get to see it in our real data). Like before, we will call $\vec{\mathbf x}_i$ the random latent positions for the nodes of our network. In this case, each $\vec {\mathbf x}_i$ is sampled independently and identically from the inner-product distribution $F$ described above. The latent-position matrix is the matrix-valued random variable $\mathbf X$ whose entries are the latent vectors $\vec {\mathbf x}_i$, for each of the $n$ nodes.
The model for edges of the *a posteriori* RDPG can be described by conditioning on this unobserved latent-position matrix. We write down that, conditioned on $\vec {\mathbf x}_i = \vec x$ and $\vec {\mathbf x}_j = \vec y$, that if $j > i$, then $\mathbf a_{ij}$ is sampled independently from a $Bern(\vec x^\top \vec y)$ distribution. As before, if $i < j$, $\mathbf a_{ji} = \mathbf a_{ij}$ (the network is *undirected*), and $\mathbf a_{ii} = 0$ (the network is *loopless*). If $\mathbf A$ is the adjacency matrix for an *a posteriori* RDPG with parameter $F$, we write that $\mathbf A \sim RDPG_n(F)$.
#### Probability
The probability for the *a posteriori* RDPG is fairly complicated. This is because, like the *a posteriori* SBM, we do not actually get to see the latent position matrix $\mathbf X$, so we need to use *integration* to obtain an expression for the probability. Here, we are concerned with realizations of $\mathbf X$. Remember that $\mathbf X$ is just a matrix whose rows are $\vec {\mathbf x}_i$, each of which individually have have the distribution $F$; e.g., $\vec{\mathbf x}_i \sim F$ independently. For simplicity, we will assume that $F$ is a disrete distribution on $\mathcal X_K$. This makes the logic of what is going on below much simpler since the notation gets less complicated, but does not detract from the generalizability of the result (the only difference is that sums would be replaced by multivariate integrals, and probability mass functions replaced by probability density functions).
We will let $p$ denote the probability mass function (p.m.f.) of this discrete distribution function $F$. The strategy will be to use the independence assumption, followed by integration over the relevant rows of $\mathbf X$:
\begin{align*}
\mathbb P_\theta(A) &= \mathbb P_\theta(\mathbf A = A) \\
&= \prod_{j > i} \mathbb P(\mathbf a_{ij} = a_{ij}), \;\;\;\;\textrm{Independence Assumption} \\
\mathbb P(\mathbf a_{ij} = a_{ij})&= \sum_{\vec x \in \mathcal X_K}\sum_{\vec y \in \mathcal X_K}\mathbb P(\mathbf a_{ij} = a_{ij}, \vec{\mathbf x}_i = \vec x, \vec{\mathbf x}_j = \vec y),\;\;\;\;\textrm{integration over }\vec {\mathbf x}_i \textrm{ and }\vec {\mathbf x}_j
\end{align*}
Next, we will simplify this expression a little bit more, using the definition of a conditional probability like we did before for the SBM:
\begin{align*}
\\
\mathbb P(\mathbf a_{ij} = a_{ij}, \vec{\mathbf x}_i = \vec x, \vec{\mathbf x}_j = \vec y) &= \mathbb P(\mathbf a_{ij} = a_{ij}| \vec{\mathbf x}_i = \vec x, \vec{\mathbf x}_j = \vec y) \mathbb P(\vec{\mathbf x}_i = \vec x, \vec{\mathbf x}_j = \vec y)
\end{align*}
Further, remember that if $\mathbf a$ and $\mathbf b$ are independent, then $\mathbb P(\mathbf a = a, \mathbf b = b) = \mathbb P(\mathbf a = a)\mathbb P(\mathbf b = b)$. Using that $\vec x_i$ and $\vec x_j$ are independent, by definition:
\begin{align*}
\mathbb P(\vec{\mathbf x}_i = \vec x, \vec{\mathbf x}_j = \vec y) &= \mathbb P(\vec{\mathbf x}_i = \vec x) \mathbb P(\vec{\mathbf x}_j = \vec y)
\end{align*}
Which means that:
\begin{align*}
\mathbb P(\mathbf a_{ij} = a_{ij}, \vec{\mathbf x}_i = \vec x, \vec{\mathbf x}_j = \vec y) &= \mathbb P(\mathbf a_{ij} = a_{ij} | \vec{\mathbf x}_i = \vec x, \vec{\mathbf x}_j = \vec y)\mathbb P(\vec{\mathbf x}_i = \vec x) \mathbb P(\vec{\mathbf x}_j = \vec y)
\end{align*}
Finally, we that conditional on $\vec{\mathbf x}_i = \vec x_i$ and $\vec{\mathbf x}_j = \vec x_j$, $\mathbf a_{ij}$ is $Bern(\vec x_i^\top \vec x_j)$. This means that in terms of our probability matrix, each entry $p_{ij} = \vec x_i^\top \vec x_j$. Therefore:
\begin{align*}
\mathbb P(\mathbf a_{ij} = a_{ij}| \vec{\mathbf x}_i = \vec x, \vec{\mathbf x}_j = \vec y) &= (\vec x^\top \vec y)^{a_{ij}}(1 - \vec x^\top\vec y)^{1 - a_{ij}}
\end{align*}
This implies that:
\begin{align*}
\mathbb P(\mathbf a_{ij} = a_{ij}, \vec{\mathbf x}_i = \vec x, \vec{\mathbf x}_j = \vec y) &= (\vec x^\top \vec y)^{a_{ij}}(1 - \vec x^\top\vec y)^{1 - a_{ij}}\mathbb P(\vec{\mathbf x}_i = \vec x) \mathbb P(\vec{\mathbf x}_j = \vec y)
\end{align*}
So our complete expression for the probability is:
\begin{align*}
\mathbb P_\theta(A) &= \prod_{j > i}\sum_{\vec x \in \mathcal X_K}\sum_{\vec y \in \mathcal X_K} (\vec x^\top \vec y)^{a_{ij}}(1 - \vec x^\top\vec y)^{1 - a_{ij}}\mathbb P(\vec{\mathbf x}_i = \vec x) \mathbb P(\vec{\mathbf x}_j = \vec y)
\end{align*}
## Generalized Random Dot Product Graph (GRDPG)
The Generalized Random Dot Product Graph, or GRDPG, is the most general random network model we will consider in this book. Note that for the RDPG, the probability matrix $P$ had entries $p_{ij} = \vec x_i^\top \vec x_j$. What about $p_{ji}$? Well, $p_{ji} = \vec x_j^\top \vec x_i$, which is exactly the same as $p_{ij}$! This means that even if we were to consider a directed RDPG, the probabilities that can be captured are *always* going to be symmetric. The generalized random dot product graph, or GRDPG, relaxes this assumption. This is achieved by using *two* latent positin matrices, $X$ and $Y$, and letting $P = X Y^\top$. Now, the entries $p_{ij} = \vec x_i^\top \vec y_j$, but $p_{ji} = \vec x_j^\top \vec y_i$, which might be different.
### *A Priori* GRDPG
The *a priori* GRDPG is a GRDPG in which we know *a priori* the latent position matrices $X$ and $Y$. The *a priori* GRDPG has the following parameters:
| Parameter | Space | Description |
| --- | --- | --- |
| $X$ | $ \mathbb R^{n \times d}$ | The matrix of left latent positions for each node $n$. |
| $Y$ | $ \mathbb R^{n \times d}$ | The matrix of right latent positions for each node $n$. |
$X$ and $Y$ behave nearly the same as the latent position matrix $X$ for the *a priori* RDPG, with the exception that they will be called the **left latent position matrix** and the **right latent position matrix** respectively. Further, the vectors $\vec x_i$ will be the left latent positions, and $\vec y_i$ will be the right latent positions, for a given node $i$, for each node $i=1,...,n$.
What is the generative model for the *a priori* GRDPG? As we discussed above, given $X$ and $Y$, for all $j \neq i$, $\mathbf a_{ij} \sim Bern(\vec x_i^\top \vec y_j)$ independently. If we consider only loopless networks, $\mathbf a_{ij} = 0$. If $\mathbf A$ is an *a priori* GRDPG with left and right latent position matrices $X$ and $Y$, we write that $\mathbf A \sim GRDPG_n(X, Y)$.
### *A Posteriori* GRDPG
The *A Posteriori* GRDPG is very similar to the *a posteriori* RDPG. We have two parameters:
| Parameter | Space | Description |
| --- | --- | --- |
| F | inner-product distributions | A distribution which governs the left latent positions. |
| G | inner-product distributions | A distribution which governs the right latent positions. |
Here, we treat the left and right latent position matrices as latent variable matrices, like we did for *a posteriori* RDPG. That is, the left latent positions are sampled independently and identically from $F$, and the right latent positions $\vec y_i$ are sampled independently and identically from $G$.
The model for edges of the *a posteriori* RDPG can be described by conditioning on the unobserved left and right latent-position matrices. We write down that, conditioned on $\vec {\mathbf x}_i = \vec x$ and $\vec {\mathbf y}_j = \vec y$, that if $j \neq i$, then $\mathbf a_{ij}$ is sampled independently from a $Bern(\vec x^\top \vec y)$ distribution. As before, assuming the network is loopless, $\mathbf a_{ii} = 0$. If $\mathbf A$ is the adjacency matrix for an *a posteriori* RDPG with parameter $F$, we write that $\mathbf A \sim GRDPG_n(F, G)$.
## Inhomogeneous Erdös-Rényi (IER)
In the preceding models, we typically made assumptions about how we could characterize the edge-existence probabilities using fewer than $\binom n 2$ different probabilities (one for each edge). The reason for this is that in general, $n$ is usually relatively large, so attempting to actually learn $\binom n 2$ different probabilities is not, in general, going to be very feasible (it is *never* feasible when we have a single network, since a single network only one observation for each independent edge). Further, it is relatively difficult to ask questions for which assuming edges share *nothing* in common (even if they don't share the same probabilities, there may be properties underlying the probabilities, such as the *latent positions* that we saw above with the RDPG, that we might still want to characterize) is actually favorable.
Nonetheless, the most general model for an independent-edge random network is known as the Inhomogeneous Erdös-Rényi (IER) Random Network. An IER Random Network is characterized by the following parameters:
| Parameter | Space | Description |
| --- | --- | --- |
| $P$ | [0,1]$^{n \times n}$ | The edge probability matrix. |
The probability matrix $P$ is an $n \times n$ matrix, where each entry $p_{ij}$ is a probability (a value between $0$ and $1$). Further, if we restrict ourselves to the case of simple networks like we have done so far, $P$ will also be symmetric ($p_{ij} = p_{ji}$ for all $i$ and $j$). The generative model is similar to the preceding models we have seen: given the $(i, j)$ entry of $P$, denoted $p_{ij}$, the edges $\mathbf a_{ij}$ are independent $Bern(p_{ij})$, for any $j > i$. Further, $\mathbf a_{ii} = 0$ for all $i$ (the network is *loopless*), and $\mathbf a_{ji} = \mathbf a_{ij}$ (the network is *undirected*). If $\mathbf A$ is the adjacency maatrix for an IER network with probability matarix $P$, we write that $\mathbf A \sim IER_n(P)$.
It is worth noting that *all* of the preceding models we have discussed so far are special cases of the IER model. This means that, for instance, if we were to consider only the probability matrices where all of the entries are the same, we could represent the ER models. Similarly, if we were to only to consider the probability matrices $P$ where $P = XX^\top$, we could represent any RDPG.
The IER Random Network can be thought of as the limit of Stochastic Block Models, as the number of communities equals the number of nodes in the network. Stated another way, an SBM Random Network where each node is in its own community is equivalent to an IER Random Network. Under this formulation, note that the block matarix for such an SBM, $B$, would have $n \times n$ unique entries. Taking $P$ to be this block matrix shows that the IER is a limiting case of SBMs.
### Probability
The probability for a network which is IER is very straightforward. We use the independence assumption, and the p.m.f. of a Bernoulli-distributed random-variable $\mathbf a_{ij}$:
\begin{align*}
\mathbb P_\theta(A) &= \mathbb P(\mathbf A = A) \\
&= \prod_{j > i}p_{ij}^{a_{ij}}(1 - p_{ij})^{1 - a_{ij}}
\end{align*}
|
github_jupyter
|
import seaborn as sns
import numpy as np
from math import comb
n = np.arange(2, 51)
logAn = np.array([comb(ni, 2) for ni in n])*np.log10(2)
ax = sns.lineplot(x=n, y=logAn)
ax.set_title("")
ax.set_xlabel("Number of Nodes")
ax.set_ylabel("Number of Possible Graphs $|A_n|$ (log scale)")
ax.set_yticks([50, 100, 150, 200, 250, 300, 350])
ax.set_yticklabels(["$10^{{{pow:d}}}$".format(pow=d) for d in [50, 100, 150, 200, 250, 300, 350]])
ax;
### Probability
What is the probability for realizations of Erdös-Rényi networks? Remember that for Independent-edge graphs, that the probability can be written:
\begin{align*}
\mathbb P_{\theta}(A) &= \prod_{j > i} \mathbb P_\theta(\mathbf{a}_{ij} = a_{ij})
\end{align*}
Next, we recall that by assumption of the ER model, that the probability matrix $P = (p)$, or that $p_{ij} = p$ for all $i,j$. Therefore:
\begin{align*}
\mathbb P_\theta(A) &= \prod_{j > i} p^{a_{ij}}(1 - p)^{1 - a_{ij}} \\
&= p^{\sum_{j > i} a_{ij}} \cdot (1 - p)^{\binom{n}{2} - \sum_{j > i}a_{ij}} \\
&= p^{m} \cdot (1 - p)^{\binom{n}{2} - m}
\end{align*}
This means that the probability $\mathbb P_\theta(A)$ is a function *only* of the number of edges $m = \sum_{j > i}a_{ij}$ in the network represented by adjacency matrix $A$. The equivalence class on the Erdös-Rényi networks are the sets:
\begin{align*}
E_{i} &= \left\{A \in \mathcal A_n : m = i\right\}
\end{align*}
where $i$ index from $0$ (the minimum number of edges possible) all the way up to $n^2$ (the maximum number of edges possible). All of the relationships for equivalence classes discussed above apply to the sets $E_i$.
## Network Models for networks which aren't simple
To make the discussions a little more easy to handle, in the above descriptions and all our successive descriptions, we will describe network models for **simple networks**. To recap, networks which are simple are binary networks which are both loopless and undirected. Stated another way, simple networks are networks whose adjacency matrices are only $0$s and $1$s, they are hollow (the diagonal is entirely *0*), and symmetric (the lower and right triangles of the adjacency matrix are the *same*). What happens our networks don't quite look this way?
For now, we'll keep the assumption that the networks are binary, but we will discuss non-binary network models in a later chapter. We have three possibilities we can consider, and we will show how the "relaxations" of the assumptions change a description of a network model. A *relaxation*, in statistician speak, means that we are taking the assumptions that we had (in this case, that the networks are *simple*), and progressively making the assumptions weaker (more *relaxed*) so that they apply to other networks, too. We split these out so we can be as clear as possible about how the generative model changes with each relaxation step.
We will compare each relaxation to the statement about the generative model for the ER generative model. To recap, for a simple network, we wrote:
"Statistically, we say that for each edge $\mathbf{a}_{ij}$ for every pair of nodes where $j > i$ (in terms of the adjacency matrix, this means all of the nodes in the *upper right* triangle), that $\mathbf{a}_{ij}$ is sampled independently and identically from a *Bernoulli* distribution with probability $p$.... We assume here that the networks are undirected, which means that if an edge $\mathbf a_{ij}$ exists from node $i$ to $j$, then the edge $\mathbf a_{ji}$ also exists from node $j$ to node $i$. We also assume that the networks are loopless, which means that no edges $\mathbf a_{ii}$ can go from node $i$ to itself."
Any additional parts that are added are expressed in **<font color='green'>green</font>** font. Omitted parts are struck through with <font color='red'><strike>red</strike></font> font.
Note that these generalizations apply to *any* of the successive networks which we describe in the Network Models section, and not just the ER model!
### Binary network model which has loops, but is undirected
Here, all we want to do is relax the assumption that the network is loopless. We simply ignore the statement that edges $\mathbf a_{ii}$ cannot exist, and allow that the $\mathbf a_{ij}$ which follow a Bernoulli distribution (with some probability which depends on the network model choice) *now* applies to $j \geq i$, and not just $j > i$. We keep that an edge $\mathbf a_{ij}$ existing implies that $\mathbf a_{ji}$ also exists, which maintains the symmetry of $\mathbf A$ (and consequently, the undirectedness of the network).
Our description of the ER network changes to:
Statistically, we say that for each edge $\mathbf{a}_{ij}$ for every pair of nodes where $\mathbf{\color{green}{j \geq i}}$ (in terms of the adjacency matrix, this means all of the nodes in the *upper right* triangle **<font color='green'>and the diagonal</font>**), that $\mathbf{a}_{ij}$ is sampled independently and identically from a *Bernoulli* distribution with probability $p$.... We assume here that the networks are undirected, which means that if an edge $\mathbf a_{ij}$ exists from node $i$ to $j$, then the edge $\mathbf a_{ji}$ also exists from node $j$ to node $i$. <font color='red'><strike>We also assume that the networks are loopless, which means that no edges $\mathbf a_{ii}$ can go from node $i$ to itself.</strike></font>
### Binary network model which is loopless, but directed
Like above, we simply ignore the statement that $\mathbf a_{ji} = \mathbf a_{ij}$, which removes the symmetry of $\mathbf A$ (and consequently, removes the undirectedness of the network). We allow that the $\mathbf a_{ij}$ which follows a Bernoulli distribution now apply to $j \neq i$, and not just $j > i$. We keep that $\mathbf a_{ii} = 0$, which maintains the hollowness of $\mathbf A$ (and consequently, the undirectedness of the network).
Our description of the ER network changes to:
Statistically, we say that for each edge $\mathbf{a}_{ij}$ for every pair of nodes where $\mathbf{\color{green}{j \neq i}}$ (in terms of the adjacency matrix, this means all of the nodes <strike><font color='red'>in the *upper right* triangle</font></strike>**<font color='green'>which are not along the diagonal</font>**), that $\mathbf{a}_{ij}$ is sampled independently and identically from a *Bernoulli* distribution with probability $p$.... <font color='red'><strike>We assume here that the networks are undirected, which means that if an edge $\mathbf a_{ij}$ exists from node $i$ to $j$, then the edge $\mathbf a_{ji}$ also exists from node $j$ to node $i$.</strike></font> We also assume that the networks are loopless, which means that no edges $\mathbf a_{ii}$ can go from node $i$ to itself.
### Binary network model which is has loops and is directed
Finally, for a network which has loops and is directed, we combine the above two approaches. We ignore the statements that $\mathbf a_{ji} = \mathbf a_{ij}$, and the statement that $\mathbf a_{ii} = 0$.
Our descriptiomn of the ER network changes to:
Statistically, we say that for each edge $\mathbf{a}_{ij}$ <font color='red'><strike>where $j > i$ (in terms of the adjacency matrix, this means all of the nodes in the *upper right* triangle)</strike></font>, that $\mathbf{a}_{ij}$ is sampled independently and identically from a *Bernoulli* distribution with probability $p$, <font color='green'>for all possible combinations of nodes $j$ and $i$</font>. <font color='red'><strike>We assume here that the networks are undirected, which means that if an edge $\mathbf a_{ij}$ exists from node $i$ to $j$, then the edge $\mathbf a_{ji}$ also exists from node $j$ to node $i$. We also assume that the networks are loopless, which means that no edges $\mathbf a_{ii}$ can go from node $i$ to itself.</strike></font>
## *A Priori* Stochastic Block Model
The *a priori* SBM is an SBM in which we know ahead of time (*a priori*) which nodes are in which communities. Here, we will use the variable $K$ to denote the maximum number of different communities. The ordering of the communities does not matter; the community we call $1$ versus $2$ versus $K$ is largely a symbolic distinction (the only thing that matters is that they are *different*). The *a priori* SBM has the following parameter:
| Parameter | Space | Description |
| --- | --- | --- |
| $B$ | [0,1]$^{K \times K}$ | The block matrix, which assigns edge probabilities for pairs of communities |
To describe the *A Priori* SBM, we will designate the community each node is a part of using a vector, which has a single community assignment for each node in the network. We will call this **node assignment vector** $\vec{\tau}$, and it is a $n$-length vector (one element for each node) with elements which can take values from $1$ to $K$. In symbols, we would say that $\vec\tau \in \{1, ..., K\}^n$. What this means is that for a given element of $\vec \tau$, $\tau_i$, that $\tau_i$ is the community assignment (either $1$, $2$, so on and so forth up to $K$) for the $i^{th}$ node. If there we hahd an example where there were $2$ communities ($K = 2$) for instance, and the first two nodes are in community $1$ and the second two in community $2$, then $\vec\tau$ would be a vector which looks like:
\begin{align*}
\vec\tau &= \begin{bmatrix}1 & 1 & 2 & 2\end{bmatrix}^\top
\end{align*}
Next, let's discuss the matrix $B$, which is known as the **block matrix** of the SBM. We write down that $B \in [0, 1]^{K \times K}$, which means that the block matrix is a matrix with $K$ rows and $K$ columns. If we have a pair of nodes and know which of the $K$ communities each node is from, the block matrix tells us the probability that those two nodes are connected. If our networks are simple, the matrix $B$ is also symmetric, which means that if $b_{kk'} = p$ where $p$ is a probability, that $b_{k'k} = p$, too. The requirement of $B$ to be symmetric exists *only* if we are dealing with undirected networks.
Finally, let's think about how to write down the generative model for the *a priori* SBM. Intuitionally what we want to reflect is, if we know that node $i$ is in community $k'$ and node $j$ is in community $k$, that the $(k', k)$ entry of the block matrix is the probability that $i$ and $j$ are connected. We say that given $\tau_i = k'$ and $\tau_j = k$, $\mathbf a_{ij}$ is sampled independently from a $Bern(b_{k' k})$ distribution for all $j > i$. Note that the adjacencies $\mathbf a_{ij}$ are not *necessarily* identically distributed, because the probability depends on the community of edge $(i,j)$. If $\mathbf A$ is an *a priori* SBM network with parameter $B$, and $\vec{\tau}$ is a realization of the node-assignment vector, we write that $\mathbf A \sim SBM_{n,\vec \tau}(B)$.
### Probability
What does the probability for the *a priori* SBM look like? In our previous description, we admittedly simplified things to an extent to keep the wording down. In truth, we model the *a priori* SBM using a *latent variable* model, which means that the node assignment vector, $\vec{\pmb \tau}$, is treated as *random*. For the case of the *a priori* SBM, it just so happens that we *know* the specific value that this latent variable $\vec{\pmb \tau}$ takes, $\vec \tau$, ahead of time.
Fortunately, since $\vec \tau$ is a *parameter* of the *a priori* SBM, the probability is a bit simpler than for the *a posteriori* SBM. This is because the *a posteriori* SBM requires an integration over potential realizations of $\vec{\pmb \tau}$, whereas the *a priori* SBM does not, since we already know that $\vec{\pmb \tau}$ was realized as $\vec\tau$.
Putting these steps together gives us that:
\begin{align*}
\mathbb P_\theta(A) &= \mathbb P_{\theta}(\mathbf A = A | \vec{\pmb \tau} = \vec\tau) \\
&= \prod_{j > i} \mathbb P_\theta(\mathbf a_{ij} = a_{ij} | \vec{\pmb \tau} = \vec\tau),\;\;\;\;\textrm{Independence Assumption}
\end{align*}
Next, for the *a priori* SBM, we know that each edge $\mathbf a_{ij}$ only *actually* depends on the community assignments of nodes $i$ and $j$, so we know that $\mathbb P_{\theta}(\mathbf a_{ij} = a_{ij} | \vec{\pmb \tau} = \vec\tau) = \mathbb P(\mathbf a_{ij} = a_{ij} | \tau_i = k', \tau_j = k)$, where $k$ and $k'$ are any of the $K$ possible communities. This is because the community assignments of nodes that are not nodes $i$ and $j$ do not matter for edge $ij$, due to the independence assumption.
Next, let's think about the probability matrix $P = (p_{ij})$ for the *a priori* SBM. We know that, given that $\tau_i = k'$ and $\tau_j = k$, each adjacency $\mathbf a_{ij}$ is sampled independently and identically from a $Bern(b_{k',k})$ distribution. This means that $p_{ij} = b_{k',k}$. Completing our analysis from above:
\begin{align*}
\mathbb P_\theta(A) &= \prod_{j > i} b_{k'k}^{a_{ij}}(1 - b_{k'k})^{1 - a_{ij}} \\
&= \prod_{k,k' \in [K]}b_{k'k}^{m_{k'k}}(1 - b_{k'k})^{n_{k'k} - m_{k'k}}
\end{align*}
Where $n_{k' k}$ denotes the total number of edges possible between nodes assigned to community $k'$ and nodes assigned to community $k$. That is, $n_{k' k} = \sum_{j > i} \mathbb 1_{\tau_i = k'}\mathbb 1_{\tau_j = k}$. Further, we will use $m_{k' k}$ to denote the total number of edges observed between these two communities. That is, $m_{k' k} = \sum_{j > i}\mathbb 1_{\tau_i = k'}\mathbb 1_{\tau_j = k}a_{ij}$. Note that for a single $(k',k)$ community pair, that the probability is analogous to the probability of a realization of an ER random variable.
<!--- We can formalize this a bit more explicitly. If we let $A^{\ell k}$ be defined as the subgraph *induced* by the edges incident nodes in community $\ell$ and those in community $k$, then we can say that $A^{\ell k}$ is a directed ER random network, --->
Like the ER model, there are again equivalence classes of the sample space $\mathcal A_n$ in terms of their probability. For a two-community setting, with $\vec \tau$ and $B$ given, the equivalence classes are the sets:
\begin{align*}
E_{a,b,c}(\vec \tau, B) &= \left\{A \in \mathcal A_n : m_{11} = a, m_{21}=m_{12} = b, m_{22} = c\right\}
\end{align*}
The number of equivalence classes possible scales with the number of communities, and the manner in which nodes are assigned to communities (particularly, the number of nodes in each community).
## *A Posteriori* Stochastic Block Model
In the *a posteriori* Stochastic Block Model (SBM), we consider that node assignment to one of $K$ communities is a random variable, that we *don't* know already like te *a priori* SBM. We're going to see a funky word come up, that you're probably not familiar with, the **$K$ probability simplex**. What the heck is a probability simplex?
The intuition for a simplex is probably something you're very familiar with, but just haven't seen a word describe. Let's say I have a vector, $\vec\pi = (\pi_k)_{k \in [K]}$, which has a total of $K$ elements. $\vec\pi$ will be a vector, which indicates the *probability* that a given node is assigned to each of our $K$ communities, so we need to impose some additional constraints. Symbolically, we would say that, for all $i$, and for all $k$:
\begin{align*}
\pi_k = \mathbb P(\pmb\tau_i = k)
\end{align*}
The $\vec \pi$ we're going to use has a very special property: all of its elements are non-negative: for all $\pi_k$, $\pi_k \geq 0$. This makes sense since $\pi_k$ is being used to represent the probability of a node $i$ being in group $k$, so it certainly can't be negative. Further, there's another thing that we want our $\vec\pi$ to have: in order for each element $\pi_k$ to indicate the probability of something to be assigned to $k$, we need all of the $\pi_k$s to sum up to one. This is because of something called the Law of Total Probability. If we have $K$ total values that $\pmb \tau_i$ could take, then it is the case that:
\begin{align*}
\sum_{k=1}^K \mathbb P(\pmb \tau_i = k) = \sum_{k = 1}^K \pi_k = 1
\end{align*}
So, back to our question: how does a probability simplex fit in? Well, the $K$ probability simplex describes all of the possible values that our vector $\vec\pi$ could take! In symbols, the $K$ probability simplex is:
\begin{align*}
\left\{\vec\pi : \text{for all $k$ }\pi_k \geq 0, \sum_{k = 1}^K \pi_k = 1 \right\}
\end{align*}
So the $K$ probability simplex is just the space for all possible vectors which could indicate assignment probabilities to one of $K$ communities.
What does the probability simplex look like? Below, we take a look at the $2$-probability simplex (2-d $\vec\pi$s) and the $3$-probability simplex (3-dimensional $\vec\pi$s):
The values of $\vec\pi = (\pi)$ that are in the $K$-probability simplex are indicated by the shaded region of each figure. This comprises the $(\pi_1, \pi_2)$ pairs that fall along a diagonal line from $(0,1)$ to $(1,0)$ for the $2$-simplex, and the $(\pi_1, \pi_2, \pi_3)$ tuples that fall on the surface of the triangular shape above with nodes at $(1,0,0)$, $(0,1,0)$, and $(0,0,1)$.
This model has the following parameters:
| Parameter | Space | Description |
| --- | --- | --- |
| $\vec \pi$ | the $K$ probability simplex | The probability of a node being assigned to community $K$ |
| $B$ | [0,1]$^{K \times K}$ | The block matrix, which assigns edge probabilities for pairs of communities |
The *a posteriori* SBM is a bit more complicated than the *a priori* SBM. We will think about the *a posteriori* SBM as a variation of the *a priori* SBM, where instead of the node-assignment vector being treated as a known fixed value (the community assignments), we will treat it as *unknown*. $\vec{\pmb \tau}$ is called a *latent variable*, which means that it is a quantity that is never actually observed, but which will be useful for describing our model. In this case, $\vec{\pmb \tau}$ takes values in the space $\{1,...,K\}^n$. This means that for a given realization of $\vec{\pmb \tau}$, denoted by $\vec \tau$, that for each of the $n$ nodes in the network, we suppose that an integer value between $1$ and $K$ indicates which community a node is from. Statistically, we write that the node assignment for node $i$, denoted by $\pmb \tau_i$, is sampled independently and identically from $Categorical(\vec \pi)$. Stated another way, the vector $\vec\pi$ indicates the probability $\pi_k$ of assignment to each community $k$ in the network.
The matrix $B$ behaves exactly the same as it did with the *a posteriori* SBM. Finally, let's think about how to write down the generative model in the *a posteriori* SBM. The model for the *a posteriori* SBM is, in fact, nearly the same as for the *a priori* SBM: we still say that given $\tau_i = k'$ and $\tau_j = k$, that $\mathbf a_{ij}$ are independent $Bern(b_{k'k})$. Here, however, we also describe that $\pmb \tau_i$ are sampled independent and identically from $Categorical(\vec\pi)$, as we learned above. If $\mathbf A$ is the adjacency matrix for an *a posteriori* SBM network with parameters $\vec \pi$ and $B$, we write that $\mathbf A \sim SBM_n(\vec \pi, B)$.
### Probability
What does the probability for the *a posteriori* SBM look like? In this case, $\theta = (\vec \pi, B)$ are the parameters for the model, so the probability for a realization $A$ of $\mathbf A$ is:
\begin{align*}
\mathbb P_\theta(A) &= \mathbb P_\theta(\mathbf A = A)
\end{align*}
Next, we use the fact that the probability that $\mathbf A = A$ is, in fact, the *integration* (over realizations of $\vec{\pmb \tau}$) of the joint $(\mathbf A, \vec{\pmb \tau})$. In this case, we will let $\mathcal T = \{1,...,K\}^n$ be the space of all possible realizations that $\vec{\pmb \tau}$ could take:
\begin{align}
\mathbb P_\theta(A)&= \sum_{\vec \tau \in \mathcal T} \mathbb P_\theta(\mathbf A = A, \vec{\pmb \tau} = \vec \tau)
\end{align}
Next, remember that by definition of a conditional probability for a random variable $\mathbf x$ taking value $x$ conditioned on random variable $\mathbf y$ taking the value $y$, that $\mathbb P(\mathbf x = x | \mathbf y = y) = \frac{\mathbb P(\mathbf x = x, \mathbf y = y)}{\mathbb P(\mathbf y = y)}$. Note that by multiplying through by $\mathbf P(\mathbf y = y)$, we can see that $\mathbb P(\mathbf x = x, \mathbf y = y) = \mathbb P(\mathbf x = x| \mathbf y = y)\mathbb P(\mathbf y = y)$. Using this logic for $\mathbf A$ and $\vec{\pmb \tau}$:
\begin{align*}
\mathbb P_\theta(A) &=\sum_{\vec \tau \in \mathcal T} \mathbb P_\theta(\mathbf A = A| \vec{\pmb \tau} = \vec \tau)\mathbb P(\vec{\pmb \tau} = \vec \tau)
\end{align*}
Intuitively, for each term in the sum, we are treating $\vec{\pmb \tau}$ as taking a fixed value, $\vec\tau$, to evaluate this probability statement.
We will start by describing $\mathbb P(\vec{\pmb \tau} = \vec\tau)$. Remember that for $\vec{\pmb \tau}$, that each entry $\pmb \tau_i$ is sampled *independently and identically* from $Categorical(\vec \pi)$.The probability mass for a $Categorical(\vec \pi)$-valued random variable is $\mathbb P(\pmb \tau_i = \tau_i; \vec \pi) = \pi_{\tau_i}$. Finally, note that if we are taking the products of $n$ $\pi_{\tau_i}$ terms, that many of these values will end up being the same. Consider, for instance, if the vector $\tau = [1,2,1,2,1]$. We end up with three terms of $\pi_1$, and two terms of $\pi_2$, and it does not matter which order we multiply them in. Rather, all we need to keep track of are the counts of each $\pi$ term. Written another way, we can use the indicator that $\tau_i = k$, given by $\mathbb 1_{\tau_i = k}$, and a running counter over all of the community probability assignments $\pi_k$ to make this expression a little more sensible. We will use the symbol $n_k = \sum_{i = 1}^n \mathbb 1_{\tau_i = k}$ to denote this value, which is the number of nodes in community $k$:
\begin{align*}
\mathbb P_\theta(\vec{\pmb \tau} = \vec \tau) &= \prod_{i = 1}^n \mathbb P_\theta(\pmb \tau_i = \tau_i),\;\;\;\;\textrm{Independence Assumption} \\
&= \prod_{i = 1}^n \pi_{\tau_i} ,\;\;\;\;\textrm{p.m.f. of a Categorical R.V.}\\
&= \prod_{k = 1}^K \pi_{k}^{n_k},\;\;\;\;\textrm{Reorganizing what we are taking products of}
\end{align*}
Next, let's think about the conditional probability term, $\mathbb P_\theta(\mathbf A = A \big | \vec{\pmb \tau} = \vec \tau)$. Remember that the entries are all independent conditional on $\vec{\pmb \tau}$ taking the value $\vec\tau$. It turns out this is exactly the same result that we obtained for the *a priori* SBM:
\begin{align*}
\mathbb P_\theta(\mathbf A = A \big | \vec{\pmb \tau} = \vec \tau)
&= \prod_{k',k} b_{\ell k}^{m_{k' k}}(1 - b_{k' k})^{n_{k' k} - m_{k' k}}
\end{align*}
Combining these into the integrand gives:
\begin{align*}
\mathbb P_\theta(A) &= \sum_{\vec \tau \in \mathcal T} \mathbb P_\theta(\mathbf A = A \big | \vec{\pmb \tau} = \vec \tau) \mathbb P_\theta(\vec{\pmb \tau} = \vec \tau) \\
&= \sum_{\vec \tau \in \mathcal T} \prod_{k = 1}^K \left[\pi_k^{n_k}\cdot \prod_{k'=1}^K b_{k' k}^{m_{k' k}}(1 - b_{k' k})^{n_{k' k} - m_{k' k}}\right]
\end{align*}
Evaluating this sum explicitly proves to be relatively tedious and is a bit outside of the scope of this book, so we will omit it here.
## Degree-Corrected Stochastic Block Model (DCSBM)
Let's think back to our school example for the Stochastic Block Model. Remember, we had 100 students, each of whom could go to one of two possible schools: school one or school two. Our network had 100 nodes, representing each of the students. We said that the school for which each student attended was represented by their node assignment $\tau_i$ to one of two possible communities. The matrix $B$ was the block probaability matrix, where $b_{11}$ was the probability that students in school one were friends, $b_{22}$ was the probability that students in school two were friends, and $b_{12} = b_{21}$ was the probability that students were friends if they did not go to the same school. In this case, we said that $\mathbf A$ was an $SBM_n(\tau, B)$ random network.
When would this setup not make sense? Let's say that Alice and Bob both go to the same school, but Alice is more popular than Bob. In general since Alice is more popular than Bob, we might want to say that for any clasasmate, Alice gets an additional "popularity benefit" to her probability of being friends with the other classmate, and Bob gets an "unpopularity penalty." The problem here is that within a single community of an SBM, the SBM assumes that the **node degree** (the number of nodes each nodes is connected to) is the *same* for all nodes within a single community. This means that we would be unable to reflect this benefit/penalty system to Alice and Bob, since each student will have the same number of friends, on average. This problem is referred to as **community degree homogeneity** in a Stochastic Block Model Network. Community degree homogeneity just means that the node degree is *homogeneous*, or the same, for all nodes within a community.
To address this limitation, we turn to the Degree-Corrected Stochastic Block Model, or DCSBM. As with the Stochastic Block Model, there is both a *a priori* and *a posteriori* DCSBM.
### *A Priori* DCSBM
Like the *a priori* SBM, the *a priori* DCSBM is where we know which nodes are in which communities ahead of time. Here, we will use the variable $K$ to denote the number of different communiies. The *a priori* DCSBM has the following two parameters:
| Parameter | Space | Description |
| --- | --- | --- |
| $B$ | [0,1]$^{K \times K}$ | The block matrix, which assigns edge probabilities for pairs of communities |
| $\vec\theta$ | $\mathbb R^n_+$ | The degree correction vector, which adjusts the degree for pairs of nodes |
The latent community assignment vector $\vec{\pmb \tau}$ with a known *a priori* realization $\vec{\tau}$ and the block matrix $B$ are exactly the same for the *a priori* DCSBM as they were for the *a priori* SBM.
The vector $\vec\theta$ is the degree correction vector. Each entry $\theta_i$ is a positive scalar. $\theta_i$ defines how much more (or less) edges associated with node $i$ are connected due to their association with node $i$.
Finally, let's think about how to write down the generative model for the *a priori* DCSBM. We say that $\tau_i = k'$ and $\tau_j = k$, $\mathbf a_{ij}$ is sampled independently from a $Bern(\theta_i \theta_j b_{k'k})$ distribution for all $j > i$. As we can see, $\theta_i$ in a sense is "correcting" the probabilities of each adjacency to node $i$ to be higher, or lower, depending on the value of $\theta_i$ that that which is given by the block probabilities $b_{\ell k}$. If $\mathbf A$ is an *a priori* DCSBM network with parameters and $B$, we write that $\mathbf A \sim DCSBM_{n,\vec\tau}(\vec \theta, B)$.
#### Probability
The derivation for the probability is the same as for the *a priori* SBM, with the change that $p_{ij} = \theta_i \theta_j b_{k'k}$ instead of just $b_{k'k}$. This gives that the probability turns out to be:
\begin{align*}
\mathbb P_\theta(A) &= \prod_{j > i} \left(\theta_i \theta_j b_{k'k}\right)^{a_{ij}}\left(1 - \theta_i \theta_j b_{k'k}\right)^{1 - a_{ij}}
\end{align*}
The expression doesn't simplify much more due to the fact that the probabilities are dependent on the particular $i$ and $j$, so we can't just reduce the statement in terms of $n_{k'k}$ and $m_{k'k}$ like for the SBM.
### *A Posteriori* DCSBM
The *a posteriori* DCSBM is to the *a posteriori* SBM what the *a priori* DCSBM was to the *a priori* SBM. The changes are very minimal, so we will omit explicitly writing it all down here so we can get this section wrapped up, with the idea that the preceding section on the *a priori* DCSBM should tell you what needs to change. We will leave it as an exercise to the reader to write down a model and probability statement for realizations of the DCSBM.
## Random Dot Product Graph (RDPG)
### *A Priori* RDPG
The *a priori* Random Dot Product Graph is an RDPG in which we know *a priori* the latent position matrix $X$. The *a priori* RDPG has the following parameter:
| Parameter | Space | Description |
| --- | --- | --- |
| $X$ | $ \mathbb R^{n \times d}$ | The matrix of latent positions for each node $n$. |
$X$ is called the **latent position matrix** of the RDPG. We write that $X \in \mathbb R^{n \times d}$, which means that it is a matrix with real values, $n$ rows, and $d$ columns. We will use the notation $\vec x_i$ to refer to the $i^{th}$ row of $X$. $\vec x_i$ is referred to as the **latent position** of a node $i$. This looks something like this:
\begin{align*}
X = \begin{bmatrix}
\vec x_{1}^\top \\
\vdots \\
\vec x_n^\top
\end{bmatrix}
\end{align*}
Noting that $X$ has $d$ columns, this implies that $\vec x_i \in \mathbb R^d$, or that each node's latent position is a real-valued $d$-dimensional vector.
What is the generative model for the *a priori* RDPG? As we discussed above, given $X$, for all $j > i$, $\mathbf a_{ij} \sim Bern(\vec x_i^\top \vec x_j)$ independently. If $i < j$, $\mathbf a_{ji} = \mathbf a_{ij}$ (the network is *undirected*), and $\mathbf a_{ii} = 0$ (the network is *loopless*). If $\mathbf A$ is an *a priori* RDPG with parameter $X$, we write that $\mathbf A \sim RDPG_n(X)$.
<!-- TODO: return to add equivalence classes -->
#### Probability
Given $X$, the probability for an RDPG is relatively straightforward, as an RDPG is another Independent-Edge Random Graph. The independence assumption vastly simplifies our resulting expression. We will also use many of the results we've identified above, such as the p.m.f. of a Bernoulli random variable. Finally, we'll note that the probability matrix $P = (\vec x_i^\top \vec x_j)$, so $p_{ij} = \vec x_i^\top \vec x_j$:
\begin{align*}
\mathbb P_\theta(A) &= \mathbb P_\theta(A) \\
&= \prod_{j > i}\mathbb P(\mathbf a_{ij} = a_{ij}),\;\;\;\; \textrm{Independence Assumption} \\
&= \prod_{j > i}(\vec x_i^\top \vec x_j)^{a_{ij}}(1 - \vec x_i^\top \vec x_j)^{1 - a_{ij}},\;\;\;\; a_{ij} \sim Bern(\vec x_i^\top \vec x_j)
\end{align*}
Unfortunately, the probability equivalence classes are a bit harder to understand intuitionally here compared to the ER and SBM examples so we won't write them down here, but they still exist!
### *A Posteriori* RDPG
Like for the *a posteriori* SBM, the *a posteriori* RDPG introduces another strange set: the **intersection of the unit ball and the non-negative orthant**. Huh? This sounds like a real mouthful, but it turns out to be rather straightforward. You are probably already very familiar with a particular orthant: in two-dimensions, an orthant is called a quadrant. Basically, an orthant just extends the concept of a quadrant to spaces which might have more than $2$ dimensions. The non-negative orthant happens to be the orthant where all of the entries are non-negative. We call the **$K$-dimensional non-negative orthant** the set of points in $K$-dimensional real space, where:
\begin{align*}
\left\{\vec x \in \mathbb R^K : x_k \geq 0\text{ for all $k$}\right\}
\end{align*}
In two dimensions, this is the traditional upper-right portion of the standard coordinate axis. To give you a picture, the $2$-dimensional non-negative orthant is the blue region of the following figure:
Now, what is the unit ball? You are probably familiar with the idea of the unit ball, even if you haven't heard it called that specifically. Remember that the Euclidean norm for a point $\vec x$ which has coordinates $x_i$ for $i=1,...,K$ is given by the expression:
\begin{align*}
\left|\left|\vec x\right|\right|_2 = \sqrt{\sum_{i = 1}^K x_i^2}
\end{align*}
The Euclidean unit ball is just the set of points whose Euclidean norm is at most $1$. To be more specific, the **closed unit ball** with the Euclidean norm is the set of points:
\begin{align*}
\left\{\vec x \in \mathbb R^K :\left|\left|\vec x\right|\right|_2 \leq 1\right\}
\end{align*}
We draw the $2$-dimensional unit ball with the Euclidean norm below, where the points that make up the unit ball are shown in red:
Now what is their intersection? Remember that the intersection of two sets $A$ and $B$ is the set:
\begin{align*}
A \cap B &= \{x : x \in A, x \in B\}
\end{align*}
That is, each element must be in *both* sets to be in the intersection. The interesction of the unit ball and the non-negative orthant will be the set:
\begin{align*}
\mathcal X_K = \left\{\vec x \in \mathbb R^K :\left|\left|\vec x\right|\right|_2 \leq 1, x_k \geq 0 \textrm{ for all $k$}\right\}
\end{align*}
visually, this will be the set of points in the *overlap* of the unit ball and the non-negative orthant, which we show below in purple:
| 0.731826 | 0.994247 |
# A Detailed RBC Model Example
Consider the equilibrium conditions for a basic RBC model without labor:
\begin{align}
C_t^{-\sigma} & = \beta E_t \left[C_{t+1}^{-\sigma}(\alpha A_{t+1} K_{t+1}^{\alpha-1} + 1 - \delta)\right]\\
Y_t & = A_t K_t^{\alpha}\\
I_t & = K_{t+1} - (1-\delta)K_t\\
Y_t & = C_t + I_t\\
\log A_t & = \rho_a \log A_{t-1} + \epsilon_t
\end{align}
In the nonstochastic steady state, we have:
\begin{align}
K & = \left(\frac{\alpha A}{1/\beta+\delta-1}\right)^{\frac{1}{1-\alpha}}\\
Y & = AK^{\alpha}\\
I & = \delta K\\
C & = Y - I
\end{align}
Given values for the parameters $\beta$, $\sigma$, $\alpha$, $\delta$, and $A$, steady state values of capital, output, investment, and consumption are easily computed.
## Import requisite modules
```
# Import numpy, pandas, linearsolve, matplotlib.pyplot
import numpy as np
import pandas as pd
import linearsolve as ls
import matplotlib.pyplot as plt
plt.style.use('classic')
%matplotlib inline
```
## Initializing the model in `linearsolve`
To initialize the model, we need to first set the model's parameters. We do this by creating a Pandas Series variable called `parameters`:
```
# Input model parameters
parameters = pd.Series()
parameters['alpha'] = .35
parameters['beta'] = 0.99
parameters['delta'] = 0.025
parameters['rhoa'] = .9
parameters['sigma'] = 1.5
parameters['A'] = 1
```
Next, we need to define a function that returns the equilibrium conditions of the model. The function will take as inputs two vectors: one vector of "current" variables and another of "forward-looking" or one-period-ahead variables. The function will return an array that represents the equilibirum conditions of the model. We'll enter each equation with all variables moved to one side of the equals sign. For example, here's how we'll enter the produciton fucntion:
`production_function = technology_current*capital_current**alpha - output_curent`
Here the variable `production_function` stores the production function equation set equal to zero. We can enter the equations in almost any way we want. For example, we could also have entered the production function this way:
`production_function = 1 - output_curent/technology_current/capital_current**alpha`
One more thing to consider: the natural log in the equation describing the evolution of total factor productivity will create problems for the solution routine later on. So rewrite the equation as:
\begin{align}
A_{t+1} & = A_{t}^{\rho_a}e^{\epsilon_{t+1}}\\
\end{align}
So the complete system of equations that we enter into the program looks like:
\begin{align}
C_t^{-\sigma} & = \beta E_t \left[C_{t+1}^{-\sigma}(\alpha Y_{t+1} /K_{t+1}+ 1 - \delta)\right]\\
Y_t & = A_t K_t^{\alpha}\\
I_t & = K_{t+1} - (1-\delta)K_t\\
Y_t & = C_t + I_t\\
A_{t+1} & = A_{t}^{\rho_a}e^{\epsilon_{t+1}}
\end{align}
Now let's define the function that returns the equilibrium conditions:
```
# Define function to compute equilibrium conditions
def equilibrium_equations(variables_forward,variables_current,parameters):
# Parameters
p = parameters
# Variables
fwd = variables_forward
cur = variables_current
# Household Euler equation
euler_eqn = p.beta*fwd.c**-p.sigma*(p.alpha*fwd.y/fwd.k+1-p.delta) - cur.c**-p.sigma
# Production function
production_fuction = cur.a*cur.k**p.alpha - cur.y
# Capital evolution
capital_evolution = fwd.k - (1-p.delta)*cur.k - cur.i
# Goods market clearing
market_clearing = cur.c + cur.i - cur.y
# Exogenous technology
technology_proc = cur.a**p.rhoa- fwd.a
# Stack equilibrium conditions into a numpy array
return np.array([
euler_eqn,
production_fuction,
capital_evolution,
market_clearing,
technology_proc
])
```
Notice that inside the function we have to define the variables of the model form the elements of the input vectors `variables_forward` and `variables_current`. It is *essential* that the predetermined or state variables are ordered first.
## Initializing the model
To initialize the model, we need to specify the number of state variables in the model, the names of the endogenous varaibles in the same order used in the `equilibrium_equations` function, and the names of the exogenous shocks to the model.
```
# Initialize the model
rbc = ls.model(equations = equilibrium_equations,
n_states=2,
var_names=['a','k','c','y','i'],
shock_names=['e_a','e_k'],
parameters=parameters)
```
The solution routine solves the model as if there were a separate exogenous shock for each state variable and that's why I initialized the model with two exogenous shocks `e_a` and `e_k` even though the RBC model only has one exogenous shock.
## Steady state
Next, we need to compute the nonstochastic steady state of the model. The `.compute_ss()` method can be used to compute the steady state numerically. The method's default is to use scipy's `fsolve()` function, but other scipy root-finding functions can be used: `root`, `broyden1`, and `broyden2`. The optional argument `options` lets the user pass keywords directly to the optimization function. Check out the documentation for Scipy's nonlinear solvers here: http://docs.scipy.org/doc/scipy/reference/optimize.html
```
# Compute the steady state numerically
guess = [1,1,1,1,1]
rbc.compute_ss(guess)
print(rbc.ss)
```
Note that the steady state is returned as a Pandas Series. Alternatively, you could compute the steady state directly and then sent the `rbc.ss` attribute:
```
# Steady state solution
p = parameters
K = (p.alpha*p.A/(1/p.beta+p.delta-1))**(1/(1-p.alpha))
C = p.A*K**p.alpha - p.delta*K
Y = p.A*K**p.alpha
I = Y - C
rbc.set_ss([p.A,K,C,Y,I])
print(rbc.ss)
```
## Log-linearization and solution
Now we use the `.log_linear()` method to find the log-linear appxoximation to the model's equilibrium conditions. That is, we'll transform the nonlinear model into a linear model in which all variables are expressed as log-deviations from the steady state. Specifically, we'll compute the matrices $A$ and $B$ that satisfy:
\begin{align}
A E_t\left[ x_{t+1} \right] & = B x_t + \left[ \begin{array}{c} \epsilon_{t+1} \\ 0 \end{array} \right],
\end{align}
where the vector $x_{t}$ denotes the log deviation of the endogenous variables from their steady state values.
```
# Find the log-linear approximation around the non-stochastic steady state
rbc.log_linear_approximation()
print('The matrix A:\n\n',np.around(rbc.a,4),'\n\n')
print('The matrix B:\n\n',np.around(rbc.b,4))
```
Finally, we need to obtain the *solution* to the log-linearized model. The solution is a pair of matrices $F$ and $P$ that specify:
1. The current values of the non-state variables $u_{t}$ as a linear function of the previous values of the state variables $s_t$.
1. The future values of the state variables $s_{t+1}$ as a linear function of the previous values of the state variables $s_t$ and the future realisation of the exogenous shock process $\epsilon_{t+1}$.
\begin{align}
u_t & = Fs_t\\
s_{t+1} & = Ps_t + \epsilon_{t+1}.
\end{align}
We use the `.klein()` method to find the solution.
```
# Solve the model
rbc.solve_klein(rbc.a,rbc.b)
# Display the output
print('The matrix F:\n\n',np.around(rbc.f,4),'\n\n')
print('The matrix P:\n\n',np.around(rbc.p,4))
```
## Impulse responses
One the model is solved, use the `.impulse()` method to compute impulse responses to exogenous shocks to the state. The method creates the `.irs` attribute which is a dictionary with keys equal to the names of the exogenous shocks and the values are Pandas DataFrames with the computed impulse respones. You can supply your own values for the shocks, but the default is 0.01 for each exogenous shock.
```
# Compute impulse responses and plot
rbc.impulse(T=41,t0=1,shocks=None,percent=True)
print('Impulse responses to a 0.01 unit shock to A:\n\n',rbc.irs['e_a'].head())
```
Plotting is easy.
```
rbc.irs['e_a'][['a','k','c','y','i']].plot(lw='5',alpha=0.5,grid=True).legend(loc='upper right',ncol=2)
rbc.irs['e_a'][['e_a','a']].plot(lw='5',alpha=0.5,grid=True).legend(loc='upper right',ncol=2)
```
## Stochastic simulation
Creating a stochastic simulation of the model is straightforward with the `.stoch_sim()` method. In the following example, I create a 151 period (including t=0) simulation by first simulating the model for 251 periods and then dropping the first 100 values. The variance of the shock to $A_t$ is set to 0.001 and the variance of the shock to $K_t$ is set to zero because there is not capital shock in the model. The seed for the numpy random number generator is set to 0.
```
rbc.stoch_sim(T=121,drop_first=100,cov_mat=np.array([[0.00763**2,0],[0,0]]),seed=0,percent=True)
rbc.simulated[['k','c','y','i']].plot(lw='5',alpha=0.5,grid=True).legend(loc='upper right',ncol=4)
rbc.simulated[['a']].plot(lw='5',alpha=0.5,grid=True).legend(ncol=4)
rbc.simulated[['e_a','e_k']].plot(lw='5',alpha=0.5,grid=True).legend(ncol=4)
```
|
github_jupyter
|
# Import numpy, pandas, linearsolve, matplotlib.pyplot
import numpy as np
import pandas as pd
import linearsolve as ls
import matplotlib.pyplot as plt
plt.style.use('classic')
%matplotlib inline
# Input model parameters
parameters = pd.Series()
parameters['alpha'] = .35
parameters['beta'] = 0.99
parameters['delta'] = 0.025
parameters['rhoa'] = .9
parameters['sigma'] = 1.5
parameters['A'] = 1
# Define function to compute equilibrium conditions
def equilibrium_equations(variables_forward,variables_current,parameters):
# Parameters
p = parameters
# Variables
fwd = variables_forward
cur = variables_current
# Household Euler equation
euler_eqn = p.beta*fwd.c**-p.sigma*(p.alpha*fwd.y/fwd.k+1-p.delta) - cur.c**-p.sigma
# Production function
production_fuction = cur.a*cur.k**p.alpha - cur.y
# Capital evolution
capital_evolution = fwd.k - (1-p.delta)*cur.k - cur.i
# Goods market clearing
market_clearing = cur.c + cur.i - cur.y
# Exogenous technology
technology_proc = cur.a**p.rhoa- fwd.a
# Stack equilibrium conditions into a numpy array
return np.array([
euler_eqn,
production_fuction,
capital_evolution,
market_clearing,
technology_proc
])
# Initialize the model
rbc = ls.model(equations = equilibrium_equations,
n_states=2,
var_names=['a','k','c','y','i'],
shock_names=['e_a','e_k'],
parameters=parameters)
# Compute the steady state numerically
guess = [1,1,1,1,1]
rbc.compute_ss(guess)
print(rbc.ss)
# Steady state solution
p = parameters
K = (p.alpha*p.A/(1/p.beta+p.delta-1))**(1/(1-p.alpha))
C = p.A*K**p.alpha - p.delta*K
Y = p.A*K**p.alpha
I = Y - C
rbc.set_ss([p.A,K,C,Y,I])
print(rbc.ss)
# Find the log-linear approximation around the non-stochastic steady state
rbc.log_linear_approximation()
print('The matrix A:\n\n',np.around(rbc.a,4),'\n\n')
print('The matrix B:\n\n',np.around(rbc.b,4))
# Solve the model
rbc.solve_klein(rbc.a,rbc.b)
# Display the output
print('The matrix F:\n\n',np.around(rbc.f,4),'\n\n')
print('The matrix P:\n\n',np.around(rbc.p,4))
# Compute impulse responses and plot
rbc.impulse(T=41,t0=1,shocks=None,percent=True)
print('Impulse responses to a 0.01 unit shock to A:\n\n',rbc.irs['e_a'].head())
rbc.irs['e_a'][['a','k','c','y','i']].plot(lw='5',alpha=0.5,grid=True).legend(loc='upper right',ncol=2)
rbc.irs['e_a'][['e_a','a']].plot(lw='5',alpha=0.5,grid=True).legend(loc='upper right',ncol=2)
rbc.stoch_sim(T=121,drop_first=100,cov_mat=np.array([[0.00763**2,0],[0,0]]),seed=0,percent=True)
rbc.simulated[['k','c','y','i']].plot(lw='5',alpha=0.5,grid=True).legend(loc='upper right',ncol=4)
rbc.simulated[['a']].plot(lw='5',alpha=0.5,grid=True).legend(ncol=4)
rbc.simulated[['e_a','e_k']].plot(lw='5',alpha=0.5,grid=True).legend(ncol=4)
| 0.697197 | 0.989368 |
# Python | day 3 | collections & if/else
### Exercise 1.
1. Create a variable called **num_bridge** with a value of 15.
```
num_bridge = 15
```
2. Create a variable called **name_street** with the value "Recoletos".
```
name_street = 'Recoletos'
```
3. Create a variable called **personal_taste** which should be `True` if you like the beach better or `False` if you rather choose mountain.
```
personal_taste = False
```
4. Create a variable called **nothing** like follows: `nothing = None`. `None`doesn't represent any value, something you should keep in mind if you do `if nothing:` then `nothing` acts as a boolean `False`.
```
nothing = None
```
5. Create a variable called **address_list**. This should be a list which contains the following: first element **name_street** and second element **num_bridge**.
```
address_list = [name_street, num_bridge]
address_list
```
6. Create a variable called **sleep_hours** that should contains, as a string, the hours you've slept today.
```
sleep_hours = '7'
```
7. Create a list called **info_list** containing every variable you've just created in the order you have done it.
```python
len(info_list) == 6
True
```
**IMPORTANT: you will use `info_list` at some point next week**
```
info_list = [num_bridge, name_street, personal_taste, nothing, address_list, sleep_hours]
info_list
```
8. Run the following cell in order to delete every variable you've just created and keep **info_list** only.
Print **info_list** once you've done it to check the list still exists.
```
#run this
del num_bridge
del name_street
del personal_taste
del nothing
del address_list
del sleep_hours
print(info_list)
```
### Exercise 2.
You'll need `info_list` in order to continue with this exercise. Remember don't use any variable except `info_list`.
Print the following. Don't use functions and/or loops.
1. Add the number of The Bridge and the hours you slept last night.
```
info_list[0] + int(info_list[-1])
```
2. Concatenate the number of The Bridge, the name of the street, your personal taste and the hours you slept. All of them should be separated by `" --> "`.
```
print(info_list[1] + '\t' + str(info_list[2]) + '\t' + info_list[-1])
```
3. Access to the list which is at the fourth position of **info_list** and print the result of:
- The concatenation of both elements contained in the list found in fourth position of **info_list**.
- The multiplication of the lenght of the list and the lenght of the street's name.
If at the fourth position of **info_list** you don't find a list, go back to Exercise 1, please.
```
print(info_list[4][0] + str(info_list[4][1]))
print(len(info_list) * len(info_list[4][0]))
```
### Exercise 3.
1. Get user input using input(“Enter your age: ”). If user is 18 or older, give feedback: You are old enough to drive. If below 18 give feedback to wait for the missing amount of years.
```
age = int(input("Enter your age: "))
if age >= 18:
print("You are old enough to drive")
else: print("Wait for another", 18 - age, "years")
```
2. Compare the values of **my_age** and **your_age** using if/else. Who is older (me or you)? Use input(“Enter your age: ”) to get **your_age** as input. **my_age** is always your age, cause you are the one coding the program.
```
my_age = 26
your_age = int(input("Enter your age: "))
if my_age > your_age:
print("I'm older by " + str(abs(my_age - your_age)) + " years")
elif my_age < your_age:
print("You are older by " + str(abs(my_age - your_age)) + " years")
elif my_age == your_age:
print("We are the same age")
else:
None
```
3. Get two numbers from the user using input prompt. If a is greater than b return "a is greater than b", if a is less b return "a is smaller than b", else "a is equal to b".
```
a = int(input("Enter value: "))
b = int(input("Enter value: "))
if a > b:
print("a is greater than b")
elif b > a:
print("b is greater than a")
else:
print("a is equal to b")
```
### Bonus track.
Learn about **for loop**
Watch this video: https://www.youtube.com/watch?v=wxds6MAtUQ0
Read this: https://www.w3schools.com/python/python_for_loops.asp
You may need this to understand what the f*** you're doing: http://www.pythontutor.com/visualize.html#mode=edit
Declare four variables:
- A string variable containing your surname.
- Two integers variables: one, with the number of the street in which you live, and two, the age of the classmate sitting on your right.
If you are on the right edge of the table, look for your classmate on your left.
If you are on remote, choose one of your mates who are on remote too.
- A list containing the variables you have just declared.
```
my_name = "Juan"
num_street = 149
age_mate = 32
var_list = [my_name, num_street, age_mate]
```
Using a **for loop** print every element of the list you've just created.
```
for i in range(0, len(var_list)):
print(var_list[i])
```
Using a **for loop** and a **if** make sure you only print the element in the list which is a string
(Your surname).
```
for i in range(0, len(var_list)):
if type(var_list[i]) == type("string"):
print(var_list[i])
```
**Well done!**
### Bonus track of the bonus track.
The following list contains some fruits:
`fruits = ['banana', 'orange', 'mango', 'lemon']`
If a fruit doesn't exist in the list add the fruit to the list and print the modified list. If the fruit exists print('That fruit already exist in the list')
```
fruits = ['banana', 'orange', 'mango', 'lemon']
in_fruit = input("Enter fruit name: ")
if in_fruit in fruit_in_list:
fruits.append(in_fruit)
print(fruits)
else:
print("That fruit already exists in the list")
```

|
github_jupyter
|
num_bridge = 15
name_street = 'Recoletos'
personal_taste = False
nothing = None
address_list = [name_street, num_bridge]
address_list
sleep_hours = '7'
len(info_list) == 6
True
info_list = [num_bridge, name_street, personal_taste, nothing, address_list, sleep_hours]
info_list
#run this
del num_bridge
del name_street
del personal_taste
del nothing
del address_list
del sleep_hours
print(info_list)
info_list[0] + int(info_list[-1])
print(info_list[1] + '\t' + str(info_list[2]) + '\t' + info_list[-1])
print(info_list[4][0] + str(info_list[4][1]))
print(len(info_list) * len(info_list[4][0]))
age = int(input("Enter your age: "))
if age >= 18:
print("You are old enough to drive")
else: print("Wait for another", 18 - age, "years")
my_age = 26
your_age = int(input("Enter your age: "))
if my_age > your_age:
print("I'm older by " + str(abs(my_age - your_age)) + " years")
elif my_age < your_age:
print("You are older by " + str(abs(my_age - your_age)) + " years")
elif my_age == your_age:
print("We are the same age")
else:
None
a = int(input("Enter value: "))
b = int(input("Enter value: "))
if a > b:
print("a is greater than b")
elif b > a:
print("b is greater than a")
else:
print("a is equal to b")
my_name = "Juan"
num_street = 149
age_mate = 32
var_list = [my_name, num_street, age_mate]
for i in range(0, len(var_list)):
print(var_list[i])
for i in range(0, len(var_list)):
if type(var_list[i]) == type("string"):
print(var_list[i])
fruits = ['banana', 'orange', 'mango', 'lemon']
in_fruit = input("Enter fruit name: ")
if in_fruit in fruit_in_list:
fruits.append(in_fruit)
print(fruits)
else:
print("That fruit already exists in the list")
| 0.062689 | 0.922622 |
## Discretisation with Decision Trees
Discretisation is the process of transforming continuous variables into discrete variables by creating a set of contiguous intervals that spans the range of the variable's values. **Supervised discretisation** methods use target information to create the contiguous bins or intervals. Several supervised discretisation methods have been described, see for example the article [Discretisation: An Enabling technique](http://www.public.asu.edu/~huanliu/papers/dmkd02.pdf) for a summary.
However, I have only seen the discretisation using decision trees being used both in Data Science competitions and business settings:
Discretisation using trees was implemented by the winning solution of the KDD 2009 cup: "Winning the KDD Cup Orange Challenge with Ensemble Selection" (http://www.mtome.com/Publications/CiML/CiML-v3-book.pdf).
It is also used in a peer to peer lending company in the UK. See this [blog](https://blog.zopa.com/2017/07/20/tips-honing-logistic-regression-models/) for details of the benefit of using discretisation using decision trees.
Discretisation with Decision Trees consists of using a decision tree to identify the optimal splitting points that would determine the bins or contiguous intervals:
- First, it trains a decision tree of limited depth (2, 3 or 4) using the variable we want to discretise to predict the target.
- The original variable values are then replaced by the probability returned by the tree. The probability is the same for all the observations within a single bin, thus replacing by the probability is equivalent to grouping the observations within the cut-off decided by the decision tree.
### Advantages
- The probabilistic predictions returned decision tree are monotonically related to the target.
- The new bins show decreased entropy, this is the observations within each bucket / bin are more similar to themselves than to those of other buckets / bins.
- The tree finds the bins automatically
### Disadvantages
- It may cause over-fitting
- More importantly, some tuning of tree parameters need to be done to obtain the optimal splits (e.g., depth, minimum number of samples in one partition, maximum number of partitions, and a minimum information gain). This it can be time consuming.
Below, I will demonstrate how to perform discretisation with decision trees using the Titanic dataset.
### Titanic dataset
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
% matplotlib inline
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier, export_graphviz
from sklearn.model_selection import cross_val_score
# load the numerical variables of the Titanic Dataset
data = pd.read_csv('titanic.csv', usecols = ['Age', 'Fare', 'Survived'])
data.head()
```
#### Important:
The tree should be built using the training dataset, and then used to replace the same feature in the testing dataset, to avoid over-fitting.
```
# Let's separate into train and test set
X_train, X_test, y_train, y_test = train_test_split(data[['Age', 'Fare', 'Survived']],
data.Survived, test_size=0.3,
random_state=0)
X_train.shape, X_test.shape
```
### Remove Missing Data
The variable Age contains missing data, that I will fill by extracting a random sample of the variable.
```
def impute_na(data, variable):
df = data.copy()
# random sampling
df[variable+'_random'] = df[variable]
# extract the random sample to fill the na
random_sample = X_train[variable].dropna().sample(df[variable].isnull().sum(), random_state=0)
# pandas needs to have the same index in order to merge datasets
random_sample.index = df[df[variable].isnull()].index
df.loc[df[variable].isnull(), variable+'_random'] = random_sample
return df[variable+'_random']
X_train['Age'] = impute_na(data, 'Age')
X_test['Age'] = impute_na(data, 'Age')
```
### Age
```
X_train.head()
# example: build Classification tree using Age to predict Survived
tree_model = DecisionTreeClassifier(max_depth=2)
tree_model.fit(X_train.Age.to_frame(), X_train.Survived)
X_train['Age_tree'] = tree_model.predict_proba(X_train.Age.to_frame())[:,1]
X_train.head(10)
X_train.Age_tree.unique()
```
A tree of depth 2, makes 2 splits, therefore generating 4 buckets, that is why we see 4 different probabilities in the output above.
```
# monotonic relationship with target
fig = plt.figure()
fig = X_train.groupby(['Age_tree'])['Survived'].mean().plot()
fig.set_title('Monotonic relationship between discretised Age and target')
fig.set_ylabel('Survived')
# number of passengers per probabilistic bucket / bin
X_train.groupby(['Age_tree'])['Survived'].count().plot.bar()
# median age within each bucket originated by the tree
X_train.groupby(['Age_tree'])['Age'].median().plot.bar()
# let's see the Age limits buckets generated by the tree
# by capturing the minimum and maximum age per each probability bucket,
# we get an idea of the bucket cut-offs
pd.concat( [X_train.groupby(['Age_tree'])['Age'].min(),
X_train.groupby(['Age_tree'])['Age'].max()], axis=1)
```
Thus, the decision tree generated the buckets: 0-11, 12-15, 16-63 and 46-80, with probabilities of survival of .51, .81, .37 and .1 respectively.
### Tree visualisation
```
# we can go ahead and visualise the tree by saving the model to a file, and opening that file in the below indicated link
with open("tree_model.txt", "w") as f:
f = export_graphviz(tree_model, out_file=f)
#http://webgraphviz.com
# this is what you should see if you do what is described in the previous cell
# the plot indicates the age cut-offs at each node, and also the number of samples at each node, and
# the gini
from IPython.display import Image
from IPython.core.display import HTML
PATH = "tree_visualisation.png"
Image(filename = PATH , width=1000, height=1000)
```
### Select the optimal depth
As I mentioned earlier, there are a number of parameters that you could optimise to obtain the best bin split using decision trees. Below I will optimise the tree depth for a demonstration. But remember that you could also optimise the remaining parameters of the decision tree. Visit [sklearn website](http://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html#sklearn.tree.DecisionTreeClassifier) to see which other parameters can be optimised.
```
# I will build trees of different depths, and I will calculate the roc-auc determined for the variable and
# the target for each tree
# I will then choose the depth that generates the best roc-auc
score_ls = [] # here I will store the roc auc
score_std_ls = [] # here I will store the standard deviation of the roc_auc
for tree_depth in [1,2,3,4]:
# call the model
tree_model = DecisionTreeClassifier(max_depth=tree_depth)
# train the model using 3 fold cross validation
scores = cross_val_score(tree_model, X_train.Age.to_frame(), y_train, cv=3, scoring='roc_auc')
score_ls.append(np.mean(scores))
score_std_ls.append(np.std(scores))
temp = pd.concat([pd.Series([1,2,3,4]), pd.Series(score_ls), pd.Series(score_std_ls)], axis=1)
temp.columns = ['depth', 'roc_auc_mean', 'roc_auc_std']
temp
```
We obtain the best roc-auc using depths of 1 or 2. I will select depth of 2 to proceed.
### Transform the feature using tree
```
tree_model = DecisionTreeClassifier(max_depth=2)
tree_model.fit(X_train.Age.to_frame(), X_train.Survived)
X_train['Age_tree'] = tree_model.predict_proba(X_train.Age.to_frame())[:,1]
X_test['Age_tree'] = tree_model.predict_proba(X_test.Age.to_frame())[:,1]
```
We are now ready to use those pre-processed Age features in machine learning algorithms. Why don't you go ahead and test them?
```
# let's inspect the transformed variables in train set
X_train.head()
# let's inspect the transformed variables in test set
X_test.head()
# and the unique values of each bin (train)
X_train.Age_tree.unique()
# and the unique values of each bin (test)
X_test.Age_tree.unique()
```
### Fare
### Select the optimal depth
Let's repeat the exercise with the variable Fare. Remember that fare was highly skewed and therefore would benefit from engineering to spread the information more evenly.
```
score_ls = []
score_std_ls = []
for tree_depth in [1,2,3,4]:
tree_model = DecisionTreeClassifier(max_depth=tree_depth)
scores = cross_val_score(tree_model, X_train.Fare.to_frame(), y_train, cv=3, scoring='roc_auc')
score_ls.append(np.mean(scores))
score_std_ls.append(np.std(scores))
temp = pd.concat([pd.Series([1,2,3,4]), pd.Series(score_ls), pd.Series(score_std_ls)], axis=1)
temp.columns = ['depth', 'roc_auc_mean', 'roc_auc_std']
temp
```
In this case, the best split roc_auc is obtained with a tree of depth 2, thus I will choose this one to proceed.
```
# train the decision tree and engineer Fare in train and test set
tree_model = DecisionTreeClassifier(max_depth=2)
tree_model.fit(X_train.Fare.to_frame(), X_train.Survived)
X_train['Fare_tree'] = tree_model.predict_proba(X_train.Fare.to_frame())[:,1]
X_test['Fare_tree'] = tree_model.predict_proba(X_test.Fare.to_frame())[:,1]
X_train['Fare_tree'].unique()
X_test['Fare_tree'].unique()
# let's see what are the Fare cut-offs within each bin
pd.concat( [X_train.groupby(['Fare_tree'])['Fare'].min(),
X_train.groupby(['Fare_tree'])['Fare'].max()], axis=1)
```
The tree generated 4 bins: 0-7.5, 7.5-10.5, 11-73 and > 73, each with probability of survival .1, .25, .44 and .75 respectively. Indicating that people that paid higher fares, where more likely to survive.
```
# and with this sequence of steps, we can visualise the tree
with open("tree_model.txt", "w") as f:
f = export_graphviz(tree_model, out_file=f)
#http://webgraphviz.com
PATH = "tree_fare.png"
Image(filename = PATH , width=1000, height=1000)
```
## Discretisation with trees on categorical variables
Discretisation using trees can also be used on categorical variables, to capture some insight into how well they predict the target.
```
# let's load the categorical variable cabin from the titanic
data = pd.read_csv('titanic.csv', usecols=['Cabin', 'Survived'])
# let's fill na with a new category missing
data.Cabin.fillna('Missing', inplace=True)
# and let's capture just the first letter of the cabin, ignoring the number of the cabin
data['Cabin'] = data['Cabin'].astype(str).str[0]
data.head()
data.groupby('Cabin')['Survived'].count() / np.float(len(data))
```
We can see that cabins A, F, G and T show a low frequency of passengers, so I will re group them into a 'Rare' Category to avoid over-fitting.
```
data['Cabin'] = np.where(data.Cabin.isin(['A', 'F', 'G', 'T']), 'Rare', data.Cabin)
data.groupby('Cabin')['Survived'].count() / np.float(len(data))
# lets replace the letters by numbers, without any sort of order
cabin_dict = {k:i for i, k in enumerate(data.Cabin.unique(), 0)}
data.loc[:, 'Cabin'] = data.loc[:, 'Cabin'].map(cabin_dict)
data.head()
# let's inspect how the new cabin looks like
data.Cabin.unique()
```
### Optimise depth
```
# Let's separate into train and test set
X_train, X_test, y_train, y_test = train_test_split(data[['Cabin', 'Survived']], data.Survived, test_size=0.3,
random_state=0)
X_train.shape, X_test.shape
# and now we optimise a tree split on it as we did for numerical variables
score_ls = []
score_std_ls = []
for tree_depth in [1,2,3,4]:
tree_model = DecisionTreeClassifier(max_depth=tree_depth)
scores = cross_val_score(tree_model, X_train.Cabin.to_frame(), y_train, cv=3, scoring='roc_auc')
score_ls.append(np.mean(scores))
score_std_ls.append(np.std(scores))
temp = pd.concat([pd.Series([1,2,3,4]), pd.Series(score_ls), pd.Series(score_std_ls)], axis=1)
temp.columns = ['depth', 'roc_auc_mean', 'roc_auc_std']
temp
# I will proceed with depth = 2
tree_model = DecisionTreeClassifier(max_depth=2)
tree_model.fit(X_train.Cabin.to_frame(), X_train.Survived)
X_train['Cabin_tree'] = tree_model.predict_proba(X_train.Cabin.to_frame())[:,1]
X_test['Cabin_tree'] = tree_model.predict_proba(X_test.Cabin.to_frame())[:,1]
# the output creates 3 bins instead of 4, why is that?
X_train.Cabin_tree.unique()
# the output creates 3 bins instead of 4, why is that?
X_test.Cabin_tree.unique()
with open("tree_model.txt", "w") as f:
f = export_graphviz(tree_model, out_file=f)
#http://webgraphviz.com
PATH = "tree_cabin.png"
Image(filename = PATH , width=1000, height=1000)
```
As we can see from the plot, we only obtain 3 bins, because the first split resulted already in a final node on the left.
```
# let's see what are the Fare cut-offs within each bin
pd.concat( [X_train.groupby(['Cabin_tree'])['Cabin'].min(),
X_train.groupby(['Cabin_tree'])['Cabin'].max()], axis=1)
```
The decision tree has found these buckets as the optimal ones: Cabins 0, 1-3 and 4-5, with probability of survival .3, .6 and .73 respectively.
Amazing, no?
**That is all for this demonstration. I hope you enjoyed the notebook, and see you in the next one.**
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
% matplotlib inline
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier, export_graphviz
from sklearn.model_selection import cross_val_score
# load the numerical variables of the Titanic Dataset
data = pd.read_csv('titanic.csv', usecols = ['Age', 'Fare', 'Survived'])
data.head()
# Let's separate into train and test set
X_train, X_test, y_train, y_test = train_test_split(data[['Age', 'Fare', 'Survived']],
data.Survived, test_size=0.3,
random_state=0)
X_train.shape, X_test.shape
def impute_na(data, variable):
df = data.copy()
# random sampling
df[variable+'_random'] = df[variable]
# extract the random sample to fill the na
random_sample = X_train[variable].dropna().sample(df[variable].isnull().sum(), random_state=0)
# pandas needs to have the same index in order to merge datasets
random_sample.index = df[df[variable].isnull()].index
df.loc[df[variable].isnull(), variable+'_random'] = random_sample
return df[variable+'_random']
X_train['Age'] = impute_na(data, 'Age')
X_test['Age'] = impute_na(data, 'Age')
X_train.head()
# example: build Classification tree using Age to predict Survived
tree_model = DecisionTreeClassifier(max_depth=2)
tree_model.fit(X_train.Age.to_frame(), X_train.Survived)
X_train['Age_tree'] = tree_model.predict_proba(X_train.Age.to_frame())[:,1]
X_train.head(10)
X_train.Age_tree.unique()
# monotonic relationship with target
fig = plt.figure()
fig = X_train.groupby(['Age_tree'])['Survived'].mean().plot()
fig.set_title('Monotonic relationship between discretised Age and target')
fig.set_ylabel('Survived')
# number of passengers per probabilistic bucket / bin
X_train.groupby(['Age_tree'])['Survived'].count().plot.bar()
# median age within each bucket originated by the tree
X_train.groupby(['Age_tree'])['Age'].median().plot.bar()
# let's see the Age limits buckets generated by the tree
# by capturing the minimum and maximum age per each probability bucket,
# we get an idea of the bucket cut-offs
pd.concat( [X_train.groupby(['Age_tree'])['Age'].min(),
X_train.groupby(['Age_tree'])['Age'].max()], axis=1)
# we can go ahead and visualise the tree by saving the model to a file, and opening that file in the below indicated link
with open("tree_model.txt", "w") as f:
f = export_graphviz(tree_model, out_file=f)
#http://webgraphviz.com
# this is what you should see if you do what is described in the previous cell
# the plot indicates the age cut-offs at each node, and also the number of samples at each node, and
# the gini
from IPython.display import Image
from IPython.core.display import HTML
PATH = "tree_visualisation.png"
Image(filename = PATH , width=1000, height=1000)
# I will build trees of different depths, and I will calculate the roc-auc determined for the variable and
# the target for each tree
# I will then choose the depth that generates the best roc-auc
score_ls = [] # here I will store the roc auc
score_std_ls = [] # here I will store the standard deviation of the roc_auc
for tree_depth in [1,2,3,4]:
# call the model
tree_model = DecisionTreeClassifier(max_depth=tree_depth)
# train the model using 3 fold cross validation
scores = cross_val_score(tree_model, X_train.Age.to_frame(), y_train, cv=3, scoring='roc_auc')
score_ls.append(np.mean(scores))
score_std_ls.append(np.std(scores))
temp = pd.concat([pd.Series([1,2,3,4]), pd.Series(score_ls), pd.Series(score_std_ls)], axis=1)
temp.columns = ['depth', 'roc_auc_mean', 'roc_auc_std']
temp
tree_model = DecisionTreeClassifier(max_depth=2)
tree_model.fit(X_train.Age.to_frame(), X_train.Survived)
X_train['Age_tree'] = tree_model.predict_proba(X_train.Age.to_frame())[:,1]
X_test['Age_tree'] = tree_model.predict_proba(X_test.Age.to_frame())[:,1]
# let's inspect the transformed variables in train set
X_train.head()
# let's inspect the transformed variables in test set
X_test.head()
# and the unique values of each bin (train)
X_train.Age_tree.unique()
# and the unique values of each bin (test)
X_test.Age_tree.unique()
score_ls = []
score_std_ls = []
for tree_depth in [1,2,3,4]:
tree_model = DecisionTreeClassifier(max_depth=tree_depth)
scores = cross_val_score(tree_model, X_train.Fare.to_frame(), y_train, cv=3, scoring='roc_auc')
score_ls.append(np.mean(scores))
score_std_ls.append(np.std(scores))
temp = pd.concat([pd.Series([1,2,3,4]), pd.Series(score_ls), pd.Series(score_std_ls)], axis=1)
temp.columns = ['depth', 'roc_auc_mean', 'roc_auc_std']
temp
# train the decision tree and engineer Fare in train and test set
tree_model = DecisionTreeClassifier(max_depth=2)
tree_model.fit(X_train.Fare.to_frame(), X_train.Survived)
X_train['Fare_tree'] = tree_model.predict_proba(X_train.Fare.to_frame())[:,1]
X_test['Fare_tree'] = tree_model.predict_proba(X_test.Fare.to_frame())[:,1]
X_train['Fare_tree'].unique()
X_test['Fare_tree'].unique()
# let's see what are the Fare cut-offs within each bin
pd.concat( [X_train.groupby(['Fare_tree'])['Fare'].min(),
X_train.groupby(['Fare_tree'])['Fare'].max()], axis=1)
# and with this sequence of steps, we can visualise the tree
with open("tree_model.txt", "w") as f:
f = export_graphviz(tree_model, out_file=f)
#http://webgraphviz.com
PATH = "tree_fare.png"
Image(filename = PATH , width=1000, height=1000)
# let's load the categorical variable cabin from the titanic
data = pd.read_csv('titanic.csv', usecols=['Cabin', 'Survived'])
# let's fill na with a new category missing
data.Cabin.fillna('Missing', inplace=True)
# and let's capture just the first letter of the cabin, ignoring the number of the cabin
data['Cabin'] = data['Cabin'].astype(str).str[0]
data.head()
data.groupby('Cabin')['Survived'].count() / np.float(len(data))
data['Cabin'] = np.where(data.Cabin.isin(['A', 'F', 'G', 'T']), 'Rare', data.Cabin)
data.groupby('Cabin')['Survived'].count() / np.float(len(data))
# lets replace the letters by numbers, without any sort of order
cabin_dict = {k:i for i, k in enumerate(data.Cabin.unique(), 0)}
data.loc[:, 'Cabin'] = data.loc[:, 'Cabin'].map(cabin_dict)
data.head()
# let's inspect how the new cabin looks like
data.Cabin.unique()
# Let's separate into train and test set
X_train, X_test, y_train, y_test = train_test_split(data[['Cabin', 'Survived']], data.Survived, test_size=0.3,
random_state=0)
X_train.shape, X_test.shape
# and now we optimise a tree split on it as we did for numerical variables
score_ls = []
score_std_ls = []
for tree_depth in [1,2,3,4]:
tree_model = DecisionTreeClassifier(max_depth=tree_depth)
scores = cross_val_score(tree_model, X_train.Cabin.to_frame(), y_train, cv=3, scoring='roc_auc')
score_ls.append(np.mean(scores))
score_std_ls.append(np.std(scores))
temp = pd.concat([pd.Series([1,2,3,4]), pd.Series(score_ls), pd.Series(score_std_ls)], axis=1)
temp.columns = ['depth', 'roc_auc_mean', 'roc_auc_std']
temp
# I will proceed with depth = 2
tree_model = DecisionTreeClassifier(max_depth=2)
tree_model.fit(X_train.Cabin.to_frame(), X_train.Survived)
X_train['Cabin_tree'] = tree_model.predict_proba(X_train.Cabin.to_frame())[:,1]
X_test['Cabin_tree'] = tree_model.predict_proba(X_test.Cabin.to_frame())[:,1]
# the output creates 3 bins instead of 4, why is that?
X_train.Cabin_tree.unique()
# the output creates 3 bins instead of 4, why is that?
X_test.Cabin_tree.unique()
with open("tree_model.txt", "w") as f:
f = export_graphviz(tree_model, out_file=f)
#http://webgraphviz.com
PATH = "tree_cabin.png"
Image(filename = PATH , width=1000, height=1000)
# let's see what are the Fare cut-offs within each bin
pd.concat( [X_train.groupby(['Cabin_tree'])['Cabin'].min(),
X_train.groupby(['Cabin_tree'])['Cabin'].max()], axis=1)
| 0.627495 | 0.986416 |
# Clustering Analysis of Hearthstone Cards - visualizations
This section uses dimensionality reduction techniques such as Principal Component Analysis (PCA) and T-distributed Stochastic Neighbor Embedding (TSNE) to attempt to visualize where each card is relative to one another.
----
## Introduction
**Dimensionality reduction techniques used**
- **Principal Component Analysis (PCA)**
PCA is a dimensionality deruction technique that decomposes a multi-dimensional matrix of features into a number of arbitary axes which explain the largest portion of the samples' variance. For visualizations, PCA is generally used to decompose the features into 2 dimensions and the samples are then plotted using this 2-dimensional coordinate system.
- **T-distributed Stochastic Neighbor Embedding (TSNE)**
On the other hand, TSNE uses probability distributions to express similarities between two points. It does so in such a way that if two points, A and B, were similar, given that point A is selected, point B then has a high probability of being chosen as a neighbor. TSNE then finds the positions of the points in a 2-dimensional space that minimizes the differences between the probability distributions among the pairs.
----
### Importing packages
```
import json
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
```
## Loading data
The processed data has already been generated and saved in the notebook `hs-package-kmeans.ipynb`. Reading the CSV file `cards-decks.csv`
```
# reading the csv file, using the first column as index
cards_features = pd.read_csv("./cards-decks.csv", index_col=0)
cards_features.head()
# saving the dataframe as numpy array
cards_features_array = np.array(cards_features)
```
## Performing PCA (2 dimensions)
Reducing the dimensions of the data from 1296 dimensions to 2 most important arbitary dimensions.
```
pca = PCA(n_components=2)
cards_features_array_pca = pca.fit_transform(cards_features_array)
print("Variance explained by the top 2 axes: %0.3f, %0.3f" % tuple(pca.explained_variance_ratio_))
```
The two axes calcualted by PCA are able to explain 7.1% andd 5.4% of the variace.
```
cards_features_array_pca_df = pd.DataFrame(cards_features_array_pca, columns = ['component 1', 'component 2'], index=cards_features.index)
cards_features_array_pca_df.head()
```
## Performing TSNE (2 dimensions)
Using TSNE to reduce the dimension of the dataset to 2 dimensions.
```
cards_features_array_tsne = TSNE(n_components=2, random_state=10).fit_transform(cards_features_array)
cards_features_array_tsne_df = pd.DataFrame(cards_features_array_tsne, columns = ['component 1', 'component 2'], index=cards_features.index)
cards_features_array_tsne_df.head()
```
## Plotting the PCA/TSNE data
```
# plotting a simple scatterplot without color or partitining.
fig = plt.figure(figsize=(7, 12))
ax1 = plt.subplot(2, 1, 1)
ax1.set_xlabel('Principal Component 1', fontsize=15)
ax1.set_ylabel('Principal Component 2', fontsize=15)
ax1.set_title('2-component PCA', fontsize=20)
ax1.scatter(cards_features_array_pca_df['component 1'],
cards_features_array_pca_df['component 2'],
s = 10,
alpha = 0.5)
ax1.grid()
ax1.set(aspect='equal')
ax2 = plt.subplot(2, 1, 2)
ax2.set_xlabel('TSNE Component 1', fontsize=15)
ax2.set_ylabel('TSNE Component 2', fontsize=15)
ax2.set_title('2-component TSNE', fontsize=20)
ax2.scatter(cards_features_array_tsne_df['component 1'],
cards_features_array_tsne_df['component 2'],
s = 10,
alpha = 0.5)
ax2.grid()
ax2.set(aspect='equal')
plt.savefig("cluster-nopartition.jpg");
```
### Discussion
Both plots show one major cluster. Given this, some partitioning is needed to further make sense of the cards distribution.
## Plotting the PCA/TSNE data again
Now with color coding for each card's class (or neutral).
The colors used are:
- Warrior (red)
- Shaman (blue)
- Rogue (black)
- Paladin (yellow)
- Hunter (green)
- Druid (brown)
- Warlock (purple)
- Mage (light blue)
- Priest (white)
- Neutral (grey)
First, the cards' associated classes have to be added to the dataframe.
### Cards details from JSON
JSON file contains the names and IDs of all the cards. This JSON file will be used to obtain the list of all card IDs.
```
# reading the JSON file
refs = json.load(open("./history-of-hearthstone/refs.json"))
card_data = pd.DataFrame.from_records(refs)
# dropping cards without dbfId
card_data = card_data[~card_data['dbfId'].isna()]
# dropping cards that are not collectible
card_data = card_data[~card_data['collectible'].isna()]
```
Merging the card_data to the existing dataframes for PCA and TSNE
```
cards_features_array_tsne_df = cards_features_array_tsne_df.merge(card_data.loc[:,["name", "cardClass"]], on="name", how='left', validate='one_to_one')
cards_features_array_pca_df = cards_features_array_pca_df.merge(card_data.loc[:,["name", "cardClass"]], on="name", how='left', validate='one_to_one')
```
### Plotting
Plotting scatterplots with color and partitining.
```
# defining the classes and their colors
classes = ["WARRIOR",
"SHAMAN",
"ROGUE",
"PALADIN",
"HUNTER",
"DRUID",
"WARLOCK",
"MAGE",
"PRIEST",
"NEUTRAL"]
colors = ["red",
"blue",
"black",
"yellow",
"green",
"brown",
"purple",
"cyan",
"white",
"grey"]
# plotting
fig = plt.figure(figsize=(15, 12))
ax1 = plt.subplot(1, 2, 1)
ax1.set_xlabel('Principal Component 1', fontsize=15)
ax1.set_ylabel('Principal Component 2', fontsize=15)
ax1.set_title('2-component PCA', fontsize=20)
ax1.grid()
ax2 = plt.subplot(1, 2, 2)
ax2.set_xlabel('TSNE Component 1', fontsize=15)
ax2.set_ylabel('TSNE Component 2', fontsize=15)
ax2.set_title('2-component TSNE', fontsize=20)
ax2.grid()
for hsclass, color in zip(classes,colors):
indices_toplot_pca = cards_features_array_pca_df['cardClass'] == hsclass
indices_toplot_tsne = cards_features_array_tsne_df['cardClass'] == hsclass
ax1.scatter(cards_features_array_pca_df.loc[indices_toplot_pca, 'component 1'],
cards_features_array_pca_df.loc[indices_toplot_pca, 'component 2'],
c = color,
s = 10,
alpha = 0.5,
label = hsclass)
ax2.scatter(cards_features_array_tsne_df.loc[indices_toplot_tsne, 'component 1'],
cards_features_array_tsne_df.loc[indices_toplot_tsne, 'component 2'],
c = color,
s = 10,
alpha = 0.5,
label = hsclass)
ax1.set(aspect='equal')
ax1.legend(loc='upper right')
ax2.set(aspect='equal')
ax2.legend(loc='upper right')
ax1.annotate('Azure Drake', xy=(28, 6))
ax2.annotate('Yogg-Sauron', xy=(-85, 0))
ax2.annotate('Harrison Jones', xy=(56, -35))
ax2.annotate('Sir Finley Mrrgglton', xy=(41, -15))
ax2.annotate('Leeroy Jenkins', xy=(10, 85))
plt.savefig("cluster-partition.jpg");
cards_features_array_tsne_df.sort_values(by='component 2', ascending=True)
```
### Discussion
**PCA**
PCA shows us different "streaks" of different colors. Each card occupies different streaks in the scatter plot. This is an expected behavior since cards from different cards cannot be in the same deck. As such, using decks as features makes class cards very distinct from each other. We also see neutral cards in grey being distributed into streaks too. These cards are likely the neutral cards that synergize well with the archetypes from a particular class. Interestingly, **Azure Drake** is an outlier. This is possibly due to the fact that **Azure Drake** can be included in almost every deck. As such, it is more likely to have a non-zero under most of the features and hence is perceived to be far away from other cards in terms of euclidean distance.
**TSNE**
Instead of streaks, TSNE position each class to occupy a range of angle from the centoid, like a pizza slice. Notable outliers are **Harrison Jones** and **Sir Finley Mrrgglton**. These two cards are usually included in warrior decks and we can see that the nearest neighbors are red points, which are warrior cards.
Similarly, **Leeroy Jenkins** is positioned in the rogue's region (black) and **Yogg Sauron** is under druid's region (brown). These cards are likely to be far away from their main archetypes and classes because despite being commonly included in their respective class' decks, they are also found in other classes' decks, making them slightly different from the class cards. Yogg Sauron has mage variants. Leeroy Jenkins and Sir Finley Mrrgglton are commonly included in other aggressive decks too. Harrison Jones is a good tech card against any deck that utilizes weapons.
It is also worth looking at warlock cards' distribution in TSNE. Warlock cards are found near neutral cards. This is reflective of the nature of warlock class cards and deck archetypes where players usually include a large portion of powerful neutral cards instead of class cards.
|
github_jupyter
|
import json
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
# reading the csv file, using the first column as index
cards_features = pd.read_csv("./cards-decks.csv", index_col=0)
cards_features.head()
# saving the dataframe as numpy array
cards_features_array = np.array(cards_features)
pca = PCA(n_components=2)
cards_features_array_pca = pca.fit_transform(cards_features_array)
print("Variance explained by the top 2 axes: %0.3f, %0.3f" % tuple(pca.explained_variance_ratio_))
cards_features_array_pca_df = pd.DataFrame(cards_features_array_pca, columns = ['component 1', 'component 2'], index=cards_features.index)
cards_features_array_pca_df.head()
cards_features_array_tsne = TSNE(n_components=2, random_state=10).fit_transform(cards_features_array)
cards_features_array_tsne_df = pd.DataFrame(cards_features_array_tsne, columns = ['component 1', 'component 2'], index=cards_features.index)
cards_features_array_tsne_df.head()
# plotting a simple scatterplot without color or partitining.
fig = plt.figure(figsize=(7, 12))
ax1 = plt.subplot(2, 1, 1)
ax1.set_xlabel('Principal Component 1', fontsize=15)
ax1.set_ylabel('Principal Component 2', fontsize=15)
ax1.set_title('2-component PCA', fontsize=20)
ax1.scatter(cards_features_array_pca_df['component 1'],
cards_features_array_pca_df['component 2'],
s = 10,
alpha = 0.5)
ax1.grid()
ax1.set(aspect='equal')
ax2 = plt.subplot(2, 1, 2)
ax2.set_xlabel('TSNE Component 1', fontsize=15)
ax2.set_ylabel('TSNE Component 2', fontsize=15)
ax2.set_title('2-component TSNE', fontsize=20)
ax2.scatter(cards_features_array_tsne_df['component 1'],
cards_features_array_tsne_df['component 2'],
s = 10,
alpha = 0.5)
ax2.grid()
ax2.set(aspect='equal')
plt.savefig("cluster-nopartition.jpg");
# reading the JSON file
refs = json.load(open("./history-of-hearthstone/refs.json"))
card_data = pd.DataFrame.from_records(refs)
# dropping cards without dbfId
card_data = card_data[~card_data['dbfId'].isna()]
# dropping cards that are not collectible
card_data = card_data[~card_data['collectible'].isna()]
cards_features_array_tsne_df = cards_features_array_tsne_df.merge(card_data.loc[:,["name", "cardClass"]], on="name", how='left', validate='one_to_one')
cards_features_array_pca_df = cards_features_array_pca_df.merge(card_data.loc[:,["name", "cardClass"]], on="name", how='left', validate='one_to_one')
# defining the classes and their colors
classes = ["WARRIOR",
"SHAMAN",
"ROGUE",
"PALADIN",
"HUNTER",
"DRUID",
"WARLOCK",
"MAGE",
"PRIEST",
"NEUTRAL"]
colors = ["red",
"blue",
"black",
"yellow",
"green",
"brown",
"purple",
"cyan",
"white",
"grey"]
# plotting
fig = plt.figure(figsize=(15, 12))
ax1 = plt.subplot(1, 2, 1)
ax1.set_xlabel('Principal Component 1', fontsize=15)
ax1.set_ylabel('Principal Component 2', fontsize=15)
ax1.set_title('2-component PCA', fontsize=20)
ax1.grid()
ax2 = plt.subplot(1, 2, 2)
ax2.set_xlabel('TSNE Component 1', fontsize=15)
ax2.set_ylabel('TSNE Component 2', fontsize=15)
ax2.set_title('2-component TSNE', fontsize=20)
ax2.grid()
for hsclass, color in zip(classes,colors):
indices_toplot_pca = cards_features_array_pca_df['cardClass'] == hsclass
indices_toplot_tsne = cards_features_array_tsne_df['cardClass'] == hsclass
ax1.scatter(cards_features_array_pca_df.loc[indices_toplot_pca, 'component 1'],
cards_features_array_pca_df.loc[indices_toplot_pca, 'component 2'],
c = color,
s = 10,
alpha = 0.5,
label = hsclass)
ax2.scatter(cards_features_array_tsne_df.loc[indices_toplot_tsne, 'component 1'],
cards_features_array_tsne_df.loc[indices_toplot_tsne, 'component 2'],
c = color,
s = 10,
alpha = 0.5,
label = hsclass)
ax1.set(aspect='equal')
ax1.legend(loc='upper right')
ax2.set(aspect='equal')
ax2.legend(loc='upper right')
ax1.annotate('Azure Drake', xy=(28, 6))
ax2.annotate('Yogg-Sauron', xy=(-85, 0))
ax2.annotate('Harrison Jones', xy=(56, -35))
ax2.annotate('Sir Finley Mrrgglton', xy=(41, -15))
ax2.annotate('Leeroy Jenkins', xy=(10, 85))
plt.savefig("cluster-partition.jpg");
cards_features_array_tsne_df.sort_values(by='component 2', ascending=True)
| 0.640973 | 0.982507 |
# Sta 663 Final Project: Implementation of DCMMs {-}
#### Author: Daniel Deng, Ziyuan Zhao (Equally contributed)
### Paper used: Bayesian forecasting of many count-valued time series {-}
### Installation {-}
Use the following line to install the package
required packages:
pip install pybats
pip install matplotlib
pip install pandas
pip install numpy
pip install statsmodels
pip install scipy
This package: pip install -i https://test.pypi.org/simple/ Sta663-DCMMs==2.2.7
GitHub: https://github.com/Anluzi/Ziyuan-Daniel_DCMMs
### Abstract {-}
When it comes to sporadic data with many zero observations, special treatment is needed. This project implements a novel algorithm named Dynamic Count Mixture Models, to customize this kind of data. The model framework is throughout Bayesian and the decouple/recouple concept allows parallel fast computation, via maintaining scalability and interpretability of indiviudal series.
Key words: Bayesian framework, Dynamic Count Mixture Models, Decouple/Recouple, Scalability, Parallel computation
### Background {-}
One hallmark of forecasting sporadic count-valued data is for the model to account for many zero observations. This can be a problem for a single Poisson model, since no Poisson distribution gives such a distribution. When one takes a very small mean, it accounts for the zeros, but this will lead to almost zero probability of observing any moderately larger positive values. Dynamic Count Mixture Models consider zero and non-zero observations separately, by introducing a latent factor $z_t$, which indicates whether or not there is a sale. The details are the following:
\begin{align}
\begin{split}
&z_t \sim Ber(\pi_t) \\
&y_t | z_t =
\begin{cases}
0, &z_t = 0 \\
1 + x_t, x_t \sim Po(\mu_t), &z_t = 1
\end{cases} \\
\end{split}
\label{eq:exampleEq}
\end{align}
\begin{align}
\begin{split}
\text{model equations}
\begin{cases}
&logit(\pi_t) = F_t^0\xi_t \\
&log(\mu_t) = F_t^+\theta_t \\
\end{cases}
\end{split}
\label{eq:exampleEq}
\end{align}
$F_t^0$ and $F_t^+$ are regressors, $\xi_t$ and $\theta_t$ are state vectors for Bernoulli and Poisson respectively, which evolve separately. $y_t$ gets updated only when $z_t = 1$.
Another advantage of this model is that under its Bayesian framework, one can easily obtain the full trajectory of $y_t$ and uncertainty via direct simulation.
Compared to other existing common methods for count-valued data, such as static generalized linear models, the biggest advantage for DCMMs is its time-varying and updating feature. The coefficients in the state vector $\xi_t$ and $\theta_t$ are constantly updated with time, depending on the observation at time t $z_t$ and $y_t$.
This project will explore the details of this mixture model and implement it on both simulated data and one real data.
### Algorithm Details {-}
As the name suggests, DCMMs are a mixture model of Bernoulli and Poisson DGLMs, which are both members of a larger family: Dynamic Generalized Linear Models (DGLMs). When implementing DCMMs, one fits Bernoulli and Poisson models separately, using the data observed, while upon forecasting, predictions from Poisson model depend on the predictions of Bernoulli model, as stated in the last section above. Since DCMMs are novel usage of __Dynamic Generalized Linear Models__, below are the details of DGLMs:
#### Notions and Structure {-}
- $y_t$ denotes the time series of interest, which, in the case of this paper, is the sales. It can be binary or non-negative counts.
- At any given time t, available information is denoted by $D_t = \{y_t, D_{t-1}, I_{t-1}\}$, where $I_{t-1}$ is any relevant additional information at time $t-1$.
- *$F_t, \theta_t$* are the dynamic regression vector and state vector at time $t$, respectively.
- $\lambda_t = F_t' \theta_t$, where $\lambda_t$ is the linear predictor at time $t$. It links the parameter of interest and the linear regression via link functions,
- i.e., $\lambda_t = logit(\pi_t)$ for binomial DGLM and $\lambda_t = log(\mu_t)$ for Poisson DGLM, where $\pi_t, \mu_t$ are probability of success and mean for these precesses.
- state vector $\theta_t$ evolves via $\theta_t = G_t \theta_t + w_t$ and $w_t \sim (0, W_t)$, where $G_t$ is the known evolution matrix and $w_t$ is the stochastic innovation vector.
- $w_t$ is independent of current and past states with moments $E[w_t|D_{t-1}, I_{t-1}] = 0$ and $V[w_t|D_{t-1}, I_{t-1}] = W_t$
The __updating__ and __forecasting__ cycle from t-1 to t for a DGLM is implemented as described below, examples for Bernoulli and Poisson are given:
1. Current information is summarized in mean vector and variance matrix of the posterior state vector $\theta_{t-1} | D_{t-1}, I_{t-1} \sim [m_{t-1}, C_{t-1}]$.
2. Via the evolution equation $\theta_t = G_t \theta_t + w_t$, the implied 1-step ahead prior moments at time $t$ are $\theta_t | D_{t-1}, I_{t-1} \sim [a_t, R_t]$, with $a_t = G_tC_{t-1}G_t'$ and $R_t = G_tC_{t-1}G_t' + W_t$.
3. The time $t$ conjugate prior satisfies $E[\lambda_t|D_{t-1}, I_{t-1}] = f_t = F_t'a_t$ and $V[\lambda_t|D_{t-1}, I_{t-1}] = q_t = F_t'R_tF_t$.
- i.e.
Binomial: $y_t \sim Bin(h_t, \pi_t)$, conjugate prior: $\pi_t \sim Be(\alpha_t, \beta_t)$, with $f_t = \psi(\alpha_t) - \psi(\beta_t)$ and $q_t = \psi'(\alpha_t) + \psi'(\beta_t)$, where $\psi(x), \psi'(x)$ are digamma and trigamma functions.
Poisson: $y_t \sim Poi(\mu_t)$, conjugate prior: $\mu_t \sim Ga(\alpha_t, \beta_t)$, with $f_t = \psi(\alpha_t) - log(\beta_t)$ and $q_t = \psi'(\alpha_t)$.
4. Forecast $y_t$ 1-step ahead using the conjugacy-induced predictive distribution $p(y_t|D_{t-1}, I_{t-1})$. This can be simulated trivially.
5. Observing $y_t$, update to the posterior.
- i.e.
Binomial: conjugate posterior: $\pi_t \sim Be(\alpha_t + y_t, \beta_t + h_t - y_t)$.
Poisson: conjugate posterior $\mu_t \sim Ga(\alpha_t + y_t, \beta_t + 1)$.
6. Update posterior mean and variance of the linear predictor $\lambda_t$: $g_t = E[\lambda_t|D_t]$ and $p_t = V[\lambda_t|D_t]$
7. Linear Bayes estimation gives posterior moments $m_t = a_t + R_tF_t(g_t - f_t)/q_t$ and $C_t = R_t - R_tF_tF_t'R_t'(1 - p_t/q_t)/q_t$
This completes the time $t-1$-to-$t$ evolve-predict-update cycle.
__Random effect discount factors__ are tunning parameters for DCMMs. Details are the following:
- Applicable to any DGLMs.
- Capture additional variation.
- Extended state vector: $\theta_t = (\xi_t, \theta_{t,0}')'$ and regression vector: $F_t' = (1, F_{t,0}')'$, where $\xi_t$ is a sequence of independent, zero-mean random effects and $\theta_{t,0}',F_{t,0}'$ are the baseline state vector and regression vector. Extended linear predictor: $\lambda_t = \xi_t + \lambda_{t,0}$
- $\xi_t$ provides an additional, day-specific "shocks" to latent coefficients.
- A random effect discount factor $\rho \in (0,1]$ is used to control the level of variability injected (via a similar fashion as the other discount factors): \\
i.e. \\
$q_{t,0} = V[\lambda_{t,0}|D_{t-1},I_{t-1}]$, let $v_t = V[\xi_t|D_{t-1}, I_{t-1}] = q_{t,0}(1-\rho)/\rho$, which inflates the variation of $\lambda_t$ by $(1-\rho)/\rho$
### Optimization {-}
This algorithm is designed for parallelism, so we compare the performance with and without parallel programming.
```
import warnings
warnings.filterwarnings('ignore')
from Sta663DCMMs.Examples import real_example, sim_example
from Sta663DCMMs.Data import load_james_three, load_sim_data
from Sta663DCMMs.Comparison_examples import glm_example, dglm_example
data = load_james_three()
# discount factors
rhos = [0.9]*len(data)
%%time
for df, rho in zip(data,rhos):
real_example(df, rho)
from joblib import Parallel, delayed
%%time
Parallel(n_jobs=3)(delayed(real_example)(df, rho) for df, rho in zip(data,rhos))
```
It can be seen that with parallel computation, fitting three models costs about __one sixth__ of the one without parallelism.
### Simulated Example {-}
```
load_sim_data()
sim_example()
```
Above shows the results of DCMM on a simulated sales data, using promotion indicator and net price as covariates. The model does a perfect job in terms of 90% coverage rate. Also, the black points (reality) and the yellow points (model mean) are almost identical.
### Real Example {-}
Below is a dataset of three-pointer made by Lebron James in 2017-2018 season. _home_ is the indicator of whether or not it is a home game and _minutes_ is the minutes Lebron played in that game.
```
data = load_james_three()[2]
data
rho = 0.9 # discount factor for the model
%%time
real_example(data, rho)
```
Above shows an example of DCMM on a real dataset. It does a great job, having a 95.2% coverage rate. It can be seen that the model adapts to the data and changes its posteriors, thus predictions.
### Comparison {-}
Two competitors for DCMMs are 1. Poisson GLM and 2. Poisson Dynamic GLM
#### Poisson GLM {-}
```
%%time
glm_example(data)
```
From the performance results of a static Poisson GLM above, we can see that the 90% confidence intervals are so much wider than the ones obtained from DCMM (even though it covers all the points, it has no practical use), which indicates this model does not have great precision. The time cost is practically the same and the DCMM example, there is one more variable (median) to plot. Therefore, a static Poisson GLM is not suitable for this kind of problem, compared to DCMM.
#### Poisson DGLM {-}
```
%%time
# use the same discount factor
dglm_example(data, rho)
```
From the model performance results of Poisson DGLM, we can see that the coverage is slightly lower than DCMM, while maintaining a reasonable credible interval (slightly narrower than DCMM's). When it comes to time cost, this is perceivably shorter than DCMM, since DCMM is one Poisson DGLM on top of a Bernoulli DGLM.
Comparison summary: Bayesian Dynamic models easily beat static GLM, while DCMMs slightly outperform Poisson DGLM. Advantage of DCMMs would have be displayed more if the data are more sporadic (with more zeros).
### Discussion {-}
Since DCMMs are designed to sporadic count-valued data, naturally it does a good job for these data. Despite that it is hard to see it being generalized to other kinds of data, the core idea of it which is the mixture and hierarchical structure, can be further explored. One possible model structure is Dynamic Binary Cascade Models (DBCMs), proposed by Lindsay Berry and Mike West (2020). Details are shown below.
- $n_{r,t}$ is the number of transactions with more than r units (sales).
- For each $r = 1:d$ ($r=0$ is a trial case.), where d is a specified positive integer, $\pi_{r,t}$ is the probability that the sales for a particular transaction exceeds r, given it exceeds $r-1$.
- For each $r = 1:d$, $n_{r,t} | n_{r-1, t} \sim Bin(n_{r-1, t}, \pi_{r, t})$, which is the sequence of conditional binomial distributions.
- The conditional model of $n_{r,t}$ has the dynamic binomial logistic form: $n_{r,t} | n_{r-1, t} \sim Bin(n_{r-1, t}, \pi_{r, t})$ where $logit(\pi_{r,t}) = F^0_{r,t} \xi_{r,t}$, with known dynamic regression vector $F^0_{r,t}$ and latent state vector $\xi_{r,t}$.
- $e_t \ge 0$ is the count of excess sales (more than d items).
Comments: DBCMs are essentially cascades of Binomial DGLMs with the successor conditioned on the adjacent precedent.
When it comes to promotion on the model, there still remain a lot of work. For example, the discount factor can be time adaptive, giving the model just enough variation to account for the data, but restraining it from exploding. Also, covariates for the model can be adaptive, too, which concerns some theoretical derivations.
### Appendix {-}
Below are the example codes for Lebron's 3P data, using dcmm_analysis():
```
# ## premodeling process
# Y = data.loc[:, 'three_made'].values
# X = [data.loc[:, ['home', 'minutes']].values]
# prior_length = 4
# nsamps = 500
# forecast_start = 40
# forecast_end = len(Y)-1
# ## extract df info
# s = sorted(set([d[:4] for d in data.date]))
# ## fitting the model
# samples, mod, coef = dcmm_analysis(Y, X, prior_length=prior_length, nsamps=nsamps,
# forecast_start=forecast_start, forecast_end=forecast_end,
# mean_only=False, rho=rho, ret=['forecast', 'model', 'model_coef'])
# ## obtain the mean, median and bounds of 90% credible interval
# avg = dcmm_analysis(Y, X, prior_length=prior_length, nsamps=nsamps,
# forecast_start=forecast_start, forecast_end=forecast_end,
# mean_only=True, rho=rho, ret=['forecast'])[0]
# med = np.median(samples, axis=0)
# upper = np.quantile(samples, 0.95, axis=0)
# lower = np.quantile(samples, 0.05, axis=0)
# ## calculate coverage
# forecast_period = np.linspace(forecast_start, forecast_end, forecast_end - forecast_start + 1)
# coverage = np.logical_and(Y[40:] <= upper, Y[40:] >= lower).sum() / len(forecast_period)
# ## make the plot
# fig, ax = plt.subplots(figsize=(12, 4))
# ax.plot(forecast_period, avg, '.y', label='Mean')
# ax.plot(forecast_period, med, '.r', label='Median')
# ax.plot(forecast_period, upper, '-b', label='90% Credible Interval')
# ax.plot(forecast_period, lower, '-b', label='90% Credible Interval')
# ax.plot(Y, '.k', label="Observed")
# ax.set_title("DCMM on Lebron James' Three-pointer Made (Season" + s[0] + "-" + s[1] + ")")
# ax.set_ylabel("Three-pointer Made Per Game")
# ax.annotate("Coverage rate: " + str('%1.3f' % coverage), xy=(0, 0), xytext=(50, max(Y)))
# plt.legend()
# fig.savefig("Examples_plots/"+"James3PM-(Season" + s[0] + "-" + s[1] + ").png")
# plt.show()
```
### References {-}
Berry, L. R., P. Helman, and M. West (2020). Probabilistic forecasting of heterogeneous consumer transaction-sales
time series. International Journal of Forecasting 36, 552–569. arXiv:1808.04698. Published online Nov 25 2019.
Berry, L. R. and M. West (2019). Bayesian forecasting of many count-valued time series. Journal of Business and
Economic Statistics. arXiv:1805.05232. Published online: 25 Jun 2019.
West, M. and P. J. Harrison (1997). Bayesian Forecasting & Dynamic Models (2nd ed.). Springer Verlag.
|
github_jupyter
|
import warnings
warnings.filterwarnings('ignore')
from Sta663DCMMs.Examples import real_example, sim_example
from Sta663DCMMs.Data import load_james_three, load_sim_data
from Sta663DCMMs.Comparison_examples import glm_example, dglm_example
data = load_james_three()
# discount factors
rhos = [0.9]*len(data)
%%time
for df, rho in zip(data,rhos):
real_example(df, rho)
from joblib import Parallel, delayed
%%time
Parallel(n_jobs=3)(delayed(real_example)(df, rho) for df, rho in zip(data,rhos))
load_sim_data()
sim_example()
data = load_james_three()[2]
data
rho = 0.9 # discount factor for the model
%%time
real_example(data, rho)
%%time
glm_example(data)
%%time
# use the same discount factor
dglm_example(data, rho)
# ## premodeling process
# Y = data.loc[:, 'three_made'].values
# X = [data.loc[:, ['home', 'minutes']].values]
# prior_length = 4
# nsamps = 500
# forecast_start = 40
# forecast_end = len(Y)-1
# ## extract df info
# s = sorted(set([d[:4] for d in data.date]))
# ## fitting the model
# samples, mod, coef = dcmm_analysis(Y, X, prior_length=prior_length, nsamps=nsamps,
# forecast_start=forecast_start, forecast_end=forecast_end,
# mean_only=False, rho=rho, ret=['forecast', 'model', 'model_coef'])
# ## obtain the mean, median and bounds of 90% credible interval
# avg = dcmm_analysis(Y, X, prior_length=prior_length, nsamps=nsamps,
# forecast_start=forecast_start, forecast_end=forecast_end,
# mean_only=True, rho=rho, ret=['forecast'])[0]
# med = np.median(samples, axis=0)
# upper = np.quantile(samples, 0.95, axis=0)
# lower = np.quantile(samples, 0.05, axis=0)
# ## calculate coverage
# forecast_period = np.linspace(forecast_start, forecast_end, forecast_end - forecast_start + 1)
# coverage = np.logical_and(Y[40:] <= upper, Y[40:] >= lower).sum() / len(forecast_period)
# ## make the plot
# fig, ax = plt.subplots(figsize=(12, 4))
# ax.plot(forecast_period, avg, '.y', label='Mean')
# ax.plot(forecast_period, med, '.r', label='Median')
# ax.plot(forecast_period, upper, '-b', label='90% Credible Interval')
# ax.plot(forecast_period, lower, '-b', label='90% Credible Interval')
# ax.plot(Y, '.k', label="Observed")
# ax.set_title("DCMM on Lebron James' Three-pointer Made (Season" + s[0] + "-" + s[1] + ")")
# ax.set_ylabel("Three-pointer Made Per Game")
# ax.annotate("Coverage rate: " + str('%1.3f' % coverage), xy=(0, 0), xytext=(50, max(Y)))
# plt.legend()
# fig.savefig("Examples_plots/"+"James3PM-(Season" + s[0] + "-" + s[1] + ").png")
# plt.show()
| 0.538741 | 0.96378 |
## Geodesics in Heat implementation
This notebook implements Geodesics in Heat [[Crane et al. 2014]](https://arxiv.org/pdf/1204.6216.pdf) for triangle and tet meshes.
Compare to the C++ implementation in `experiments/geodesic_heat/main.cc`.
```
import sys
sys.path.append('..')
import mesh, differential_operators, sparse_matrices, numpy as np
from tri_mesh_viewer import TriMeshViewer as Viewer
volMesh = mesh.Mesh('../../examples/meshes/3D_microstructure.msh', degree=1)
# Choose whether to work with the tet mesh or its boundary triangle mesh.
m = volMesh
#m = volMesh.boundaryMesh()
# Choose a timestep proportional to h^2 where h is the average edge length.
# (As discussed in section 3.2.4 of the paper)
c = 4 / np.sqrt(3)
t = c * m.volume / m.numElements()
# Choose source vertex/vertices for computing distances
sourceVertices = [0]
```
We have not yet bound the sparse matrix manipulation and solver functionality of MeshFEM, so we use scipy for now:
```
import scipy, scipy.sparse, scipy.sparse.linalg
```
Backwards Euler time stepping for heat equation $\frac{\mathrm{d}}{\mathrm{d}t} = \bigtriangleup u, \, \, u|_\gamma = 1 \, \forall t$:
\begin{align}
\frac{u_t - u_0}{t} &= \bigtriangleup u_t \\
\Longrightarrow \quad M \frac{u_t - u_0}{t} &= -L u_t \quad \text{(positive FEM Laplacian discretizes $-\bigtriangleup$)} \\
\Longrightarrow \quad \underbrace{(M + t L)}_A u_t &= M u_0
\end{align}
where $\gamma$ is the domain from which we wish to compute distances (here given by `sourceVertices`)
```
L = differential_operators.laplacian(m).compressedColumn()
M = differential_operators.mass(m, lumped=False).compressedColumn()
A = L + t * M
mask = np.ones(m.numVertices(), dtype=bool)
mask[sourceVertices] = False
A_ff = A[:, mask][mask, :]
A_fc = A[:, ~mask][mask, :]
# Solve (M + t L) u = 0 with the constraint u[sourceVertices] = 1
u = np.ones(m.numVertices())
u[mask] = scipy.sparse.linalg.spsolve(A_ff, -A_fc @ np.ones(len(sourceVertices)))
# Compute the heat gradients
g = differential_operators.gradient(m, u)
# Normalize the gradients to get an approximate gradient of the distance field
X = -g / np.linalg.norm(g, axis=1)[:, np.newaxis]
```
Fit a scalar field's gradients to these normalized gradients $X$ by solving a Poisson equation:
\begin{align}
- \bigtriangleup \phi = -\nabla \cdot X \quad &\text{in } \Omega \\
\frac{\mathrm{d} \phi}{\mathrm{d} {\bf n}} = {\bf n} \cdot X \quad &\text{on } \partial \Omega \\
\phi = 0 \quad &\text{on } \gamma
\end{align}
```
divX = differential_operators.divergence(m, X)
L_ff = L[:, mask][mask, :]
heatDist = np.zeros(m.numVertices())
heatDist[mask] = scipy.sparse.linalg.spsolve(L_ff, divX[mask])
```
Visualize the approximate distance field.
```
view = Viewer(m, scalarField=heatDist)
view.show()
```
|
github_jupyter
|
import sys
sys.path.append('..')
import mesh, differential_operators, sparse_matrices, numpy as np
from tri_mesh_viewer import TriMeshViewer as Viewer
volMesh = mesh.Mesh('../../examples/meshes/3D_microstructure.msh', degree=1)
# Choose whether to work with the tet mesh or its boundary triangle mesh.
m = volMesh
#m = volMesh.boundaryMesh()
# Choose a timestep proportional to h^2 where h is the average edge length.
# (As discussed in section 3.2.4 of the paper)
c = 4 / np.sqrt(3)
t = c * m.volume / m.numElements()
# Choose source vertex/vertices for computing distances
sourceVertices = [0]
import scipy, scipy.sparse, scipy.sparse.linalg
L = differential_operators.laplacian(m).compressedColumn()
M = differential_operators.mass(m, lumped=False).compressedColumn()
A = L + t * M
mask = np.ones(m.numVertices(), dtype=bool)
mask[sourceVertices] = False
A_ff = A[:, mask][mask, :]
A_fc = A[:, ~mask][mask, :]
# Solve (M + t L) u = 0 with the constraint u[sourceVertices] = 1
u = np.ones(m.numVertices())
u[mask] = scipy.sparse.linalg.spsolve(A_ff, -A_fc @ np.ones(len(sourceVertices)))
# Compute the heat gradients
g = differential_operators.gradient(m, u)
# Normalize the gradients to get an approximate gradient of the distance field
X = -g / np.linalg.norm(g, axis=1)[:, np.newaxis]
divX = differential_operators.divergence(m, X)
L_ff = L[:, mask][mask, :]
heatDist = np.zeros(m.numVertices())
heatDist[mask] = scipy.sparse.linalg.spsolve(L_ff, divX[mask])
view = Viewer(m, scalarField=heatDist)
view.show()
| 0.542621 | 0.986891 |
# Image Standards in Medicine
```
# uncomment if you need to install dminteract
#!python -m pip install -U git+https://github.com/chapmanbe/dminteract#egg=dminteract
from dminteract.modules.m4c import *
from ipywidgets import interact, fixed
import warnings
from skimage.exposure import equalize_hist
warnings.filterwarnings('ignore')
display(question_banks["qbank4"]["dd0, qbank4"])
```
<html>
<table style="width:100%">
<tr>
<td><img src="./data/ct6.jpg" alt="3D rendering" width="128" height="128">
</td>
<td><img src="./data/cor_T2_FS.jpg" alt="T2 FSE" width="128" height="128"></td>
</tr>
<tr>
<td><img src="./data/ct_2.5mm_Std.jpg" alt="T2 FSE" width="128" height="128"></td>
<td><img src="./data/FSPGR_3D_POST.jpg" alt="T2 FSE" width="128" height="128"></td>
</tr>
</table>
</html>
## [Digital Imaging and Communications in Medicine (DICOM)](https://en.wikipedia.org/wiki/DICOM)
>"This is rather confusing." ([Introduction to DICOM](https://nipy.org/nibabel/dicom/dicom_intro.html))
DICOM is the most important image standard in medicine. It dates back to the mid 1980s and came into wide-spread use in the 1990s and 2000s when picture archiving systems (PACS) came into wide spread use. **It can be rather confusing!**
DICOM is primarily a **communication** standard, how two computers can exchange imagine data. What we are going to be exploring here is the data representation standard.
## [DICOM defines a file format](http://dicom.nema.org/dicom/2013/output/chtml/part10/chapter_7.html)
## [And a DICOM Dictionary](http://dicom.nema.org/dicom/2006/06_06pu.pdf)
### Which will of the most interest to us
Here are some small excerpts from the DICOM header for one of my images:
```
(0008, 0005) Specific Character Set CS: 'ISO_IR 100'
(0008, 0008) Image Type CS: ['ORIGINAL', 'PRIMARY', 'OTHER']
(0008, 0018) SOP Instance UID UI: 1.2.840.113619.2.312.3596.3051310.12162.1328474860.590
(0008, 0020) Study Date DA: '20120206'
```
-------------------------
```
(0009, 1002) [Suite id] UN: b'UCSD'
(0009, 1004) [Product id] UN: b'SIGNA '
(0009, 1030) [Service id] UN: b'858657MR1 '
```
-------------------------
```
(0043, 1098) [ASSET Acquisition Calibration Seri UN: Array of 54 elements
(0043, 109a) [Rx Stack Identification] UN: b'1 '
(7fe0, 0010) Pixel Data OW: Array of 524288 elements
```
Let's try to analyze these. We see that each line consists of four elements:
1. **(0008, 0005)**: This is a two-tuple of 4-digit hexadecimal numbers (note: `7fe0`).
1. The first number is the **group number**
1. The second number is the **element number**
1. **Specific Character Set**: This is the name (human readable label for the concept).
1. **CS**: This is the **value representation**, that is, how the value is represented, in this case a character string
1. **'ISO_IR 100'**: This is the actual value.
DICOM also defines a **value multiplicity** which does not seem to be grabbed by the DICOM program I'm using.
As you browse through a DICOM dictionary, you will see that similar concepts tend to be collected in the same group (same group number). This implies that group numbers have semantic meaning, a violation of good standards design (see slide 22 in Omar Bouhaddou's presentation).
However, this is an artifact of the age of the DICOM standard. Since DICOM 3.0 (1993), group numbers do not have semantic meaning:
>Although similar or related Data Elements often have the same Group Number; a Data Group does not convey any semantic meaning beginning with DICOM Version 3.0. (NEMA ["The Data Set"](http://dicom.nema.org/dicom/2013/output/chtml/part05/chapter_7.html))
Odd numbered groups are **private groups.** The motivation for private groups is
>Implementations may require communication of information that cannot be contained in Standard Data Elements. Private Data Elements are intended to be used to contain such information. Such Private Data Elements shall not change the semantics of the Information Object Definition or SOP Class Definition. (NEMA ["7.8 Private Data Elements"](http://dicom.nema.org/dicom/2013/output/chtml/part05/sect_7.8.html)
You can see in the snippets above that group `0009` is being used by the institution (UCSD) to describe itself and the machine being used to generate the images.
Private tags can be problematic as manufacturers can put all sorts of unexpected information in private tags. As an example, consider de-identification. If we wish to share images with others for research purposes, we need to remove any protected information about the patient first (e.g. patient name).
I used the [Horos](https://horosproject.org/) software package to de-identify the images before using them for this course.
In the original file, my name was present in the expected `(0010,0010)` field (as rendered by Horos):
```
PatientsName (0010,0010) CHAPMAN^BRIAN^E^
```
After de-identifcation, this field is properly blanked out (as rendered by [pydicom](https://pydicom.github.io/)):
```
(0010, 0010) Patient's Name PN: ''
```
However, the institution had put my name in a private field that was missed by the Horos de-identification algorithm (as rendered by pydicom):
```
(0033, 1013) [Patient's Name] UN: b'CHAPMAN^BRIAN^E^'
```
A more aggressive de-identification strategy might be to eliminate all private tags, but this is often problematic for images acquired with new, cutting edge technologies.
```
(0032, 1030) Reason for Study LO: "Clinical History:->new onset Trigeminal neuralgia - Lt sided Last creatinine: CREAT 1.45 1/31/2012 Last BUN: BUN 23 1/31/2012 Last GFR: GFRNON 53 1/31/2012 Last GFR: No results found for this basename: GFRAA Who To Call Providers (at the time that the order was placed): Reason:->q. Special - Trigeminal Nerve Protocol Contrast:->With *** Cr at baseline, has had contrast in the past Defer to Radiologist's Protocol for Final Order?->Yes MR Angiography:->Unspecified ICD9-CM Diagnostic code : 350.1"
```
-----------
```
(0010, 21b0) Additional Patient History LT: "Clinical History:->new onset Trigeminal neuralgia - Lt sided Last creatinine: CREAT 1.45 1/31/2012 Last BUN: BUN 23 1/31/2012 Last GFR: GFRNON 53 1/31/2012 Last GFR: No results found for this basename: GFRAA Who To Call Providers (at the time that the order was placed): Reason:->q. Special - Trigeminal Nerve Protocol Contrast:->With *** Cr at baseline, has had contrast in the past Defer to Radiologist's Protocol for Final Order?->Yes MR Angiography:->Unspecified ICD9-CM Diagnostic code : 350.1"
```
### On whom was the image acquired?
Group `0010` has multiple elements describing the patient (me)---in this case they have been de-identified---including my non-existent mulitary rank!
```
(0010, 0010) Patient's Name PN: ''
(0010, 0020) Patient ID LO: ''
(0010, 0030) Patient's Birth Date DA: ''
(0010, 0040) Patient's Sex CS: 'M'
(0010, 1010) Patient's Age AS: ''
(0010, 1030) Patient's Weight DS: "90.718"
(0010, 1080) Military Rank LO: 'ROUTINE'
```
Here is the similar section from a mammography image:
```
(0010, 0010) Patient's Name PN: 'Case1'
(0010, 0020) Patient ID LO: 'Case1'
(0010, 0030) Patient's Birth Date DA: ''
(0010, 0040) Patient's Sex CS: ''
```
Notice that weight is recorded for the MRI but not the mammogram. Patient weight is an important parameter in MRI for estimating energy deposition in the subject. Weight is not important for mammography.
### How was the image acquired
Here is an excerpt from my MRI
```
(0018, 0020) Scanning Sequence CS: 'GR'
(0018, 0021) Sequence Variant CS: 'SS'
(0018, 0022) Scan Options CS: ['FAST_GEMS', 'ACC_GEMS', 'PFF']
(0018, 0023) MR Acquisition Type CS: '3D'
(0018, 0025) Angio Flag CS: 'N'
(0018, 0050) Slice Thickness DS: "1.4"
(0018, 0080) Repetition Time DS: "6.573"
(0018, 0081) Echo Time DS: "2.2"
(0018, 0082) Inversion Time DS: "0.0"
(0018, 0083) Number of Averages DS: "1.0"
(0018, 0084) Imaging Frequency DS: "127.71572"
(0018, 0085) Imaged Nucleus SH: '1H'
(0018, 0086) Echo Number(s) IS: "1"
(0018, 0087) Magnetic Field Strength DS: "3.0"
(0018, 0088) Spacing Between Slices DS: "0.7"
(0018, 0091) Echo Train Length IS: "1"
(0018, 0093) Percent Sampling DS: "100.0"
(0018, 0094) Percent Phase Field of View DS: "100.0"
(0018, 0095) Pixel Bandwidth DS: "325.508"
(0018, 1000) Device Serial Number LO: '0000000858657MR1'
(0018, 1020) Software Versions LO: ['24', 'LX', 'MR Software release:HD16.0_V02_1131.a']
(0018, 1030) Protocol Name LO: 'Alksne Trigeminal/3'
(0018, 1088) Heart Rate IS: "0"
(0018, 1090) Cardiac Number of Images IS: "0"
(0018, 1094) Trigger Window IS: "0"
(0018, 1100) Reconstruction Diameter DS: "200.0"
(0018, 1250) Receive Coil Name SH: '8HRBRAIN'
(0018, 1310) Acquisition Matrix US: [0, 512, 512, 0]
(0018, 1312) In-plane Phase Encoding Direction CS: 'ROW'
(0018, 1314) Flip Angle DS: "40.0"
(0018, 1315) Variable Flip Angle Flag CS: 'N'
```
Most of these are data that probably only make sense to an MR physicist. For example, `GR` stands for "gradient recall." Perhaps problematically from a standards point of view, the values do not have units of measurement. In the field
```
(0018, 0087) Magnetic Field Strength DS: "3.0"
```
3.0 is 3.0 T (three Tesla) and
```
(0018, 0088) Spacing Between Slices DS: "0.7"
```
is 0.7 mm.
```
(0018, 1316) SAR DS: "1.16854"
```
Some of the instantiated fields do not have meaning for this particular sequence. For example,
```
(0018, 0082) Inversion Time DS: "0.0"
```
Here are entries for a mammogram:
```
(0018, 1030) Protocol Name LO: 'L MLO'
(0018, 1110) Distance Source to Detector DS: "700.0"
(0018, 1111) Distance Source to Patient DS: "620.0"
(0018, 1114) Estimated Radiographic Magnificatio DS: "1.073"
(0018, 1150) Exposure Time IS: "306"
(0018, 1151) X-Ray Tube Current IS: "180"
(0018, 1152) Exposure IS: "60"
(0018, 1153) Exposure in uAs IS: "60000"
(0018, 1166) Grid CS: 'NONE'
(0018, 1190) Focal Spot(s) DS: "0.3"
(0018, 1191) Anode Target Material CS: 'TUNGSTEN'
(0018, 11a0) Body Part Thickness DS: "55.0"
(0018, 11a2) Compression Force DS: "139.6735"
(0018, 1200) Date of Last Calibration DA: '19990101'
(0018, 1201) Time of Last Calibration TM: '000000.000'
(0018, 1405) Relative X-Ray Exposure IS: "494"
(0018, 1508) Positioner Type CS: 'MAMMOGRAPHIC'
(0018, 1510) Positioner Primary Angle DS: "45.1"
(0018, 5101) View Position CS: 'MLO'
(0018, 7000) Detector Conditions Nominal Flag CS: 'YES'
(0018, 7001) Detector Temperature DS: "30.18"
(0018, 7004) Detector Type CS: 'DIRECT'
(0018, 700a) Detector ID SH: 'DET00000'
(0018, 700c) Date of Last Detector Calibration DA: '19990101'
(0018, 700e) Time of Last Detector Calibration TM: '000000.000'
(0018, 701a) Detector Binning DS: [2, 2]
(0018, 7030) Field of View Origin DS: [0, 508]
(0018, 7032) Field of View Rotation DS: "0.0"
(0018, 7034) Field of View Horizontal Flip CS: 'NO'
(0018, 7050) Filter Material CS: 'ALUMINUM'
(0018, 7052) Filter Thickness Minimum DS: "0.7"
(0018, 7054) Filter Thickness Maximum DS: "0.7"
(0018, 7060) Exposure Control Mode CS: 'AUTOMATIC'
(0018, 7062) Exposure Control Mode Description LT: 'AutoFilter'
(0018, 8150) Exposure Time in uS DS: "305556.0"``
```
Here we see again that imaging parameters are lumped in group `0018` (no semantic meaning!), but they are a completely different set of element numbers. Some things to point out
* Sometimes we have explicit units ("Exposure Time in uS" micro seconds, $\mu s$), but usually not
* Lots of values are free text entries, not coded values ("ALUMINUM")
### Important Observation!
All DICOM data elements do not need to be present in the header (it would be a gigantic mess if they were). In fact, DICOM defines what is required for the header based on what kind of image is being represented. A modality like MRI is a composite of various required groups that in total represent an **information entity**.
### Exploring DICOM Images
In this section we are going to read in four distinct DICOM images. We will use these images to gain some familiarity with DICOM metadata.
```
img1 = pydicom.dcmread(os.path.join("data","I1.dcm"))
img2 = pydicom.dcmread(os.path.join("data","I2.dcm"))
img3 = pydicom.dcmread(os.path.join("data","I3.dcm"))
img4 = pydicom.dcmread(os.path.join("data","I4.dcm"))
```
### Exploration approach 1
Recalling "Concept-Based Representation" we have
* The concept (unit of thought)
* A non-semantic identifier
* A linguistic term
The non-semantic identifier is the unique concept to which the linguistic term could have synonyms or be rendered in different languages, etc.
In the DICOM standard the non-semantic identifers are the group-element tuple expressed as hexadecimal integers; this is the unique value in the concept.
Let's try browsing our images by entering different group-element tuples
* Select different images with the drop down menu.
* Type in different integers in the group and elem fields
```
_= interact(view_dicom_data, img={"img1":img1,
"img2":img2,
"img3":img3,
"img4":img4}.items(), group="0x008", elem="0x008")
```
#### Unless you are really savvy with DICOM or you have a DICOM reference nearby this feels like shooting in the dark
### Exploring by Name
In the cells below, we take a more human approach. In each cell we have a different image. You can select a different DICOM element name and then see the group-element tuple and the value.
```
_ = interact(view_dicom_data_rev,
img=fixed(img1),
item=get_rev_dict(img1))
_ = interact(view_dicom_data_rev,
img=fixed(img2),
item=get_rev_dict(img2))
_ = interact(view_dicom_data_rev,
img=fixed(img3),
item=get_rev_dict(img3))
_ = interact(view_dicom_data_rev,
img=fixed(img4),
item=get_rev_dict(img4))
```
#### Use the cells above to explore the DICOM metadata to answer the following questions
```
for q in question_banks["qbank3"].values():
display(q)
```
### Before we leave Take a look at what the images look like
```
_=interact(lambda x:imshow(equalize_hist(x.pixel_array), cmap="gray"),
x={"img1":img1,
"img2":img2,
"img3":img3,
"img4":img4}.items())
```
### Modality Standards
The DICOM standard defines data elements specific to each imaging modality. You can browse the standards for the imaging modalities examined here.
* [Mammography](http://dicom.nema.org/medical/dicom/current/output/chtml/part03/sect_C.8.31.html).
* [CT](http://dicom.nema.org/medical/dicom/current/output/chtml/part03/sect_C.8.2.html)
* [MR](http://dicom.nema.org/medical/dicom/current/output/chtml/part03/sect_C.8.3.html)
### [Move onto the next notebook](dicom_intro_pixels_voxels.ipynb)
|
github_jupyter
|
# uncomment if you need to install dminteract
#!python -m pip install -U git+https://github.com/chapmanbe/dminteract#egg=dminteract
from dminteract.modules.m4c import *
from ipywidgets import interact, fixed
import warnings
from skimage.exposure import equalize_hist
warnings.filterwarnings('ignore')
display(question_banks["qbank4"]["dd0, qbank4"])
(0008, 0005) Specific Character Set CS: 'ISO_IR 100'
(0008, 0008) Image Type CS: ['ORIGINAL', 'PRIMARY', 'OTHER']
(0008, 0018) SOP Instance UID UI: 1.2.840.113619.2.312.3596.3051310.12162.1328474860.590
(0008, 0020) Study Date DA: '20120206'
(0009, 1002) [Suite id] UN: b'UCSD'
(0009, 1004) [Product id] UN: b'SIGNA '
(0009, 1030) [Service id] UN: b'858657MR1 '
(0043, 1098) [ASSET Acquisition Calibration Seri UN: Array of 54 elements
(0043, 109a) [Rx Stack Identification] UN: b'1 '
(7fe0, 0010) Pixel Data OW: Array of 524288 elements
PatientsName (0010,0010) CHAPMAN^BRIAN^E^
(0010, 0010) Patient's Name PN: ''
(0033, 1013) [Patient's Name] UN: b'CHAPMAN^BRIAN^E^'
(0032, 1030) Reason for Study LO: "Clinical History:->new onset Trigeminal neuralgia - Lt sided Last creatinine: CREAT 1.45 1/31/2012 Last BUN: BUN 23 1/31/2012 Last GFR: GFRNON 53 1/31/2012 Last GFR: No results found for this basename: GFRAA Who To Call Providers (at the time that the order was placed): Reason:->q. Special - Trigeminal Nerve Protocol Contrast:->With *** Cr at baseline, has had contrast in the past Defer to Radiologist's Protocol for Final Order?->Yes MR Angiography:->Unspecified ICD9-CM Diagnostic code : 350.1"
(0010, 21b0) Additional Patient History LT: "Clinical History:->new onset Trigeminal neuralgia - Lt sided Last creatinine: CREAT 1.45 1/31/2012 Last BUN: BUN 23 1/31/2012 Last GFR: GFRNON 53 1/31/2012 Last GFR: No results found for this basename: GFRAA Who To Call Providers (at the time that the order was placed): Reason:->q. Special - Trigeminal Nerve Protocol Contrast:->With *** Cr at baseline, has had contrast in the past Defer to Radiologist's Protocol for Final Order?->Yes MR Angiography:->Unspecified ICD9-CM Diagnostic code : 350.1"
(0010, 0010) Patient's Name PN: ''
(0010, 0020) Patient ID LO: ''
(0010, 0030) Patient's Birth Date DA: ''
(0010, 0040) Patient's Sex CS: 'M'
(0010, 1010) Patient's Age AS: ''
(0010, 1030) Patient's Weight DS: "90.718"
(0010, 1080) Military Rank LO: 'ROUTINE'
(0010, 0010) Patient's Name PN: 'Case1'
(0010, 0020) Patient ID LO: 'Case1'
(0010, 0030) Patient's Birth Date DA: ''
(0010, 0040) Patient's Sex CS: ''
(0018, 0020) Scanning Sequence CS: 'GR'
(0018, 0021) Sequence Variant CS: 'SS'
(0018, 0022) Scan Options CS: ['FAST_GEMS', 'ACC_GEMS', 'PFF']
(0018, 0023) MR Acquisition Type CS: '3D'
(0018, 0025) Angio Flag CS: 'N'
(0018, 0050) Slice Thickness DS: "1.4"
(0018, 0080) Repetition Time DS: "6.573"
(0018, 0081) Echo Time DS: "2.2"
(0018, 0082) Inversion Time DS: "0.0"
(0018, 0083) Number of Averages DS: "1.0"
(0018, 0084) Imaging Frequency DS: "127.71572"
(0018, 0085) Imaged Nucleus SH: '1H'
(0018, 0086) Echo Number(s) IS: "1"
(0018, 0087) Magnetic Field Strength DS: "3.0"
(0018, 0088) Spacing Between Slices DS: "0.7"
(0018, 0091) Echo Train Length IS: "1"
(0018, 0093) Percent Sampling DS: "100.0"
(0018, 0094) Percent Phase Field of View DS: "100.0"
(0018, 0095) Pixel Bandwidth DS: "325.508"
(0018, 1000) Device Serial Number LO: '0000000858657MR1'
(0018, 1020) Software Versions LO: ['24', 'LX', 'MR Software release:HD16.0_V02_1131.a']
(0018, 1030) Protocol Name LO: 'Alksne Trigeminal/3'
(0018, 1088) Heart Rate IS: "0"
(0018, 1090) Cardiac Number of Images IS: "0"
(0018, 1094) Trigger Window IS: "0"
(0018, 1100) Reconstruction Diameter DS: "200.0"
(0018, 1250) Receive Coil Name SH: '8HRBRAIN'
(0018, 1310) Acquisition Matrix US: [0, 512, 512, 0]
(0018, 1312) In-plane Phase Encoding Direction CS: 'ROW'
(0018, 1314) Flip Angle DS: "40.0"
(0018, 1315) Variable Flip Angle Flag CS: 'N'
(0018, 0087) Magnetic Field Strength DS: "3.0"
(0018, 0088) Spacing Between Slices DS: "0.7"
(0018, 1316) SAR DS: "1.16854"
(0018, 0082) Inversion Time DS: "0.0"
(0018, 1030) Protocol Name LO: 'L MLO'
(0018, 1110) Distance Source to Detector DS: "700.0"
(0018, 1111) Distance Source to Patient DS: "620.0"
(0018, 1114) Estimated Radiographic Magnificatio DS: "1.073"
(0018, 1150) Exposure Time IS: "306"
(0018, 1151) X-Ray Tube Current IS: "180"
(0018, 1152) Exposure IS: "60"
(0018, 1153) Exposure in uAs IS: "60000"
(0018, 1166) Grid CS: 'NONE'
(0018, 1190) Focal Spot(s) DS: "0.3"
(0018, 1191) Anode Target Material CS: 'TUNGSTEN'
(0018, 11a0) Body Part Thickness DS: "55.0"
(0018, 11a2) Compression Force DS: "139.6735"
(0018, 1200) Date of Last Calibration DA: '19990101'
(0018, 1201) Time of Last Calibration TM: '000000.000'
(0018, 1405) Relative X-Ray Exposure IS: "494"
(0018, 1508) Positioner Type CS: 'MAMMOGRAPHIC'
(0018, 1510) Positioner Primary Angle DS: "45.1"
(0018, 5101) View Position CS: 'MLO'
(0018, 7000) Detector Conditions Nominal Flag CS: 'YES'
(0018, 7001) Detector Temperature DS: "30.18"
(0018, 7004) Detector Type CS: 'DIRECT'
(0018, 700a) Detector ID SH: 'DET00000'
(0018, 700c) Date of Last Detector Calibration DA: '19990101'
(0018, 700e) Time of Last Detector Calibration TM: '000000.000'
(0018, 701a) Detector Binning DS: [2, 2]
(0018, 7030) Field of View Origin DS: [0, 508]
(0018, 7032) Field of View Rotation DS: "0.0"
(0018, 7034) Field of View Horizontal Flip CS: 'NO'
(0018, 7050) Filter Material CS: 'ALUMINUM'
(0018, 7052) Filter Thickness Minimum DS: "0.7"
(0018, 7054) Filter Thickness Maximum DS: "0.7"
(0018, 7060) Exposure Control Mode CS: 'AUTOMATIC'
(0018, 7062) Exposure Control Mode Description LT: 'AutoFilter'
(0018, 8150) Exposure Time in uS DS: "305556.0"``
img1 = pydicom.dcmread(os.path.join("data","I1.dcm"))
img2 = pydicom.dcmread(os.path.join("data","I2.dcm"))
img3 = pydicom.dcmread(os.path.join("data","I3.dcm"))
img4 = pydicom.dcmread(os.path.join("data","I4.dcm"))
_= interact(view_dicom_data, img={"img1":img1,
"img2":img2,
"img3":img3,
"img4":img4}.items(), group="0x008", elem="0x008")
_ = interact(view_dicom_data_rev,
img=fixed(img1),
item=get_rev_dict(img1))
_ = interact(view_dicom_data_rev,
img=fixed(img2),
item=get_rev_dict(img2))
_ = interact(view_dicom_data_rev,
img=fixed(img3),
item=get_rev_dict(img3))
_ = interact(view_dicom_data_rev,
img=fixed(img4),
item=get_rev_dict(img4))
for q in question_banks["qbank3"].values():
display(q)
_=interact(lambda x:imshow(equalize_hist(x.pixel_array), cmap="gray"),
x={"img1":img1,
"img2":img2,
"img3":img3,
"img4":img4}.items())
| 0.438785 | 0.974629 |
420-A52-SF - Algorithmes d'apprentissage supervisé - Hiver 2020 - Spécialisation technique en Intelligence Artificielle - Mikaël Swawola, M.Sc.
<br/>

<br/>
**Objectif:** cette séance de travaux pratique consiste en la mise en oeuvre sous forme de code vectorisé de l'**algorithme du gradient en régression linéaire multiple**. Le jeu de données utilisé sera la version complète du jeu de données *Advertising* et devra être **mis à l'échelle**
```
%reload_ext autoreload
%autoreload 2
%matplotlib inline
```
### 0 - Chargement des bibliothèques
```
# Manipulation de données
import numpy as np
import pandas as pd
from collections import defaultdict
# Visualisation de données
import matplotlib.pyplot as plt
import seaborn as sns
# Outils divers
from tqdm.notebook import tqdm_notebook
from tqdm import tqdm
# Configuration de la visualisation
sns.set(style="darkgrid", rc={'figure.figsize':(11.7,8.27)})
```
### 1 - Lecture du jeu de données advertising
**Exercice 1-1**: à l'aide de la bibiothèques *pandas*, lire le fichier `advertising-multivariate.csv`
```
# Compléter le code ci-dessous ~ 1 ligne
df = pd.read_csv('../../data/advertising-multivariate.csv', usecols=['TV','radio','newspaper','sales'])
```
**Exercice 1-2**: à l'aide de la fonction `head()`, visualiser les premières lignes de la trame de données. Quelle sera la taille du vecteur de paramètres $\theta$ ?
```
# Compléter le code ci-dessous ~ 1 ligne
df.head()
```
### 2 - Mise à l'échelle des données
**Exercice 2**: Standardiser les données.<br/>
Note: Il n'est pas nécéssaire de standardiser la variable de sortie, mais vous pouvez le faire à des fins de simplification
```
# Compléter le code ci-dessous ~ 1 ligne
df_norm = (df - df.mean(axis=0))/df.std(axis=0)
df_norm.head()
```
### 3 - Préparation de la structure de données
**Exercice 3**: Construire la matrice des prédicteurs X sans oublier d'ajouter une colonne représentant $x_0$
```
# Compléter le code ci-dessous ~ 5 lignes
x0 = np.ones(shape=(200))
x1 = df_norm['TV'].values
x2 = df_norm['radio'].values
x3 = df_norm['newspaper'].values
X = np.array((x0,x1,x2,x3))
y = df['sales'].values.reshape(-1,1) # Nous gardons ici les valeurs non standardisées
```
<strong style='color: green'>TEST - Le code ci-dessous vous permet de tester la forme de `X`. Le `assert` ne doit pas renvoyer d'exception</strong>
```
assert X.shape == (4,200)
```
### 4 - Définition du modèle
**Exercice 4**: compléter la fonction ci-dessous représentant le modèle de régression linéaire multiple (hypothèse)
Pour rappel, le modèle de régression multiple est
$h_{\theta}(x)=\theta_{0}x_0 + \theta_{1}x_1 + \cdots + \theta_{n}x_n = \theta^TX$
```
def hypothesis(x, theta):
assert x.shape[0] == theta.shape[0]
# Compléter le code ~ 1 ligne
h = np.dot(theta.T, x)
return h
```
<strong style='color: green'>TEST - Le code ci-dessous vous permet de tester votre fonction `hypothesis`. Le `assert` ne doit pas renvoyer d'exception</strong>
```
x_test = np.array([[1,1],[3,4],[2,2],[1,-1]])
theta_test = np.array([1,2,2,4]).reshape(-1,1)
hypothesis(x_test, theta_test)
assert np.array_equal(hypothesis(x_test,theta_test), np.array([[15,9]]))
```
### 5 - Fonction de coût
**Exercice 5**: compléter la fonction ci-dessous permettant le calcul du coût (fonction de coût)
Pour rappel, la fonction de coût en régression linéaire multiple s'exprime sous la forme
$J(\theta)= \frac{1}{2m}\sum\limits_{i=1}^{m}(h_{\theta}(x^{(i)})-y^{(i)})^{2}=\frac{1}{2m}(y-X^t\theta)^T\times(y-X^t\theta)$
Remarque: comme le montre l'équation ci-dessus, il exite deux méthodes pour calculer la fonction de coût. Choisissez celle qui vous convient.<br/><em>Optionnel: faites l'autre méthode</em>
```
def cost_function(x,y, theta):
# Compléter le code ~ 1-4 lignes
error = hypothesis(x, theta) - y.T
squared_error = error**2
sse = np.sum(squared_error)
cost = (1/(2*y.shape[0])) * sse
return cost
```
<strong style='color: green'>TEST - Le code ci-dessous permet de tester la fonction `cost_function`. Celle-ci doit retourner un `numpy.float64`, c'est-à-dire un nombre et non tableau (array). Le `assert` ne doit pas renvoyer d'exception et le résultat attendu est ~ 94.92</strong>
```
theta_test = np.array([1,2,2,4]).reshape(-1,1)
cost = cost_function(X,y,theta_test)
assert type(cost) == np.float64
cost
def cost_function(x,y, theta):
# Compléter le code ~ 1-4 lignes
error = (y.reshape(-1,1) - np.dot(X.T,theta))
sse = np.dot(error.T, error)
cost = (1/(2*y.shape[0])) * sse.squeeze()
return cost
```
<strong style='color: green'>TEST - Le code ci-dessous permet de tester la fonction `cost_function`. Celle-ci doit retourner un `numpy.float64`, c'est-à-dire un nombre et non tableau (array). Le `assert` ne doit pas renvoyer d'exception et le résultat attendu est ~ 94.92</strong>
```
theta_test = np.array([1,2,2,4]).reshape(-1,1)
cost = cost_function(X,y,theta_test)
assert type(cost) == np.float64
cost
```
### 6 - Algorithme du gradient
**Exercice 6**: Compléter l'algorithme du gradient ci-dessous. Choisir le vecteur $\theta$ initial, la valeurs du **pas** ($\alpha$) et le **nombre d'itérations**. Un test de convergence ne sera pas utilisé ici.
$
\text{Répéter pendant n_iterations}
\{\\
\theta_{j}:= \theta_{j} - \alpha\frac{1}{m}\sum\limits_{i=1}^{m}(h_{\theta}(x^{(i)})-y^{(i)})\times x_{j}^{(i)}\quad\forall j
\\
\}
$
Ou sous forme vectorisée:
$
\text{Répéter pendant n_iterations}
\{\\
\theta:= \theta - \alpha\frac{1}{m}(\theta^TX-y)\times X
\\
\}
$
<strong>Vous êtes vivement encouragés à utiliser la forme vectorisée. Celle-ci est de toute façon plus simple à coder que la version non vectorisée !</strong>
```
theta = np.zeros(shape = (X.shape[0],1))
alpha = 0.000066
n_iterations = 650000
m = y.shape[0]
history = defaultdict(list)
for i in tqdm(range(0, n_iterations)):
# Compléter le code ~ 2 lignes
d_theta = np.dot((np.dot(theta.T,X) - y.T), X.T) / m
theta = theta - (alpha * d_theta.T)
# Sauvegarde des valeurs intermédiaires de theta et du coût
if i%50 == 0:
cost = cost_function(X, y, theta)
history['theta_0'].append(theta[0])
history['theta_1'].append(theta[1])
history['theta_2'].append(theta[2])
history['theta_3'].append(theta[3])
history['cost'].append(cost)
print(f'Theta = {theta}')
```
Les valeurs des paramètres $\theta_j$ devraient approcher
```[[14.0225 ]
[ 3.92908869]
[ 2.79906919]
[-0.02259517]]```
**Version (partiellement) non vectorisée**
```
theta = np.zeros(shape = (X.shape[0],1))
alpha = 0.000066
n_iterations = 650000
m = y.shape[0]
history = defaultdict(list)
for i in tqdm(range(0, n_iterations)):
d_theta = np.zeros(shape=(X.shape[0],1))
for j in range(0,X.shape[0]):
d_theta[j] = np.sum((hypothesis(X, theta) - y)*X[j,:]) / m
theta = theta - (alpha * d_theta)
# Sauvegarde des valeurs intermédiaires de theta et du coût
if i%50 == 0:
cost = cost_function(X, y, theta)
history['theta_0'].append(theta[0])
history['theta_1'].append(theta[1])
history['theta_2'].append(theta[2])
history['theta_3'].append(theta[3])
history['cost'].append(cost)
print(f'Theta = {theta}')
```
### 7 - Interprétation des paramètres
**Exercice 7**: Interpréter les paramètres obtenus
### Fin du TP
|
github_jupyter
|
%reload_ext autoreload
%autoreload 2
%matplotlib inline
# Manipulation de données
import numpy as np
import pandas as pd
from collections import defaultdict
# Visualisation de données
import matplotlib.pyplot as plt
import seaborn as sns
# Outils divers
from tqdm.notebook import tqdm_notebook
from tqdm import tqdm
# Configuration de la visualisation
sns.set(style="darkgrid", rc={'figure.figsize':(11.7,8.27)})
# Compléter le code ci-dessous ~ 1 ligne
df = pd.read_csv('../../data/advertising-multivariate.csv', usecols=['TV','radio','newspaper','sales'])
# Compléter le code ci-dessous ~ 1 ligne
df.head()
# Compléter le code ci-dessous ~ 1 ligne
df_norm = (df - df.mean(axis=0))/df.std(axis=0)
df_norm.head()
# Compléter le code ci-dessous ~ 5 lignes
x0 = np.ones(shape=(200))
x1 = df_norm['TV'].values
x2 = df_norm['radio'].values
x3 = df_norm['newspaper'].values
X = np.array((x0,x1,x2,x3))
y = df['sales'].values.reshape(-1,1) # Nous gardons ici les valeurs non standardisées
assert X.shape == (4,200)
def hypothesis(x, theta):
assert x.shape[0] == theta.shape[0]
# Compléter le code ~ 1 ligne
h = np.dot(theta.T, x)
return h
x_test = np.array([[1,1],[3,4],[2,2],[1,-1]])
theta_test = np.array([1,2,2,4]).reshape(-1,1)
hypothesis(x_test, theta_test)
assert np.array_equal(hypothesis(x_test,theta_test), np.array([[15,9]]))
def cost_function(x,y, theta):
# Compléter le code ~ 1-4 lignes
error = hypothesis(x, theta) - y.T
squared_error = error**2
sse = np.sum(squared_error)
cost = (1/(2*y.shape[0])) * sse
return cost
theta_test = np.array([1,2,2,4]).reshape(-1,1)
cost = cost_function(X,y,theta_test)
assert type(cost) == np.float64
cost
def cost_function(x,y, theta):
# Compléter le code ~ 1-4 lignes
error = (y.reshape(-1,1) - np.dot(X.T,theta))
sse = np.dot(error.T, error)
cost = (1/(2*y.shape[0])) * sse.squeeze()
return cost
theta_test = np.array([1,2,2,4]).reshape(-1,1)
cost = cost_function(X,y,theta_test)
assert type(cost) == np.float64
cost
theta = np.zeros(shape = (X.shape[0],1))
alpha = 0.000066
n_iterations = 650000
m = y.shape[0]
history = defaultdict(list)
for i in tqdm(range(0, n_iterations)):
# Compléter le code ~ 2 lignes
d_theta = np.dot((np.dot(theta.T,X) - y.T), X.T) / m
theta = theta - (alpha * d_theta.T)
# Sauvegarde des valeurs intermédiaires de theta et du coût
if i%50 == 0:
cost = cost_function(X, y, theta)
history['theta_0'].append(theta[0])
history['theta_1'].append(theta[1])
history['theta_2'].append(theta[2])
history['theta_3'].append(theta[3])
history['cost'].append(cost)
print(f'Theta = {theta}')
**Version (partiellement) non vectorisée**
| 0.42919 | 0.952486 |
This notebook suppliments the `OpenFF-benchmark-ligand-fragments-v1.0` dataset which was fragmented using fragmenter=0.7.0 (openeye only) with new molecules generated using openff-fragmenter=0.1.2 with both openeye and ambertools.
## Workflow
- First we use openeye to fragment the dataset
- Next fragment again using ambertools
- Then we combine the two datasets together using qcsubmit. This will deduplicate molecules and torsion drives but does not check for conformer duplication.
- Load the orginal dataset and add the new combined dataset to it
- Now loop over the original dataset and for any scans which are old replace the input molecules with the orginal inputs to help QCFractal deduplicate the tasks.
```
from openff.qcsubmit.factories import TorsiondriveDatasetFactory
from openff.qcsubmit import workflow_components
from openff.qcsubmit.datasets import TorsiondriveDataset, load_dataset
from openff.qcsubmit.serializers import deserialize
from openff.toolkit.topology import Molecule
from openff.toolkit.utils.toolkits import GLOBAL_TOOLKIT_REGISTRY, OpenEyeToolkitWrapper, AmberToolsToolkitWrapper
# configure the toolkit registry to only use openeye, openff-fragmenter should respect this
GLOBAL_TOOLKIT_REGISTRY.deregister_toolkit(AmberToolsToolkitWrapper())
print(GLOBAL_TOOLKIT_REGISTRY.registered_toolkits)
factory = TorsiondriveDatasetFactory()
factory.add_workflow_components(workflow_components.WBOFragmenter(keep_non_rotor_ring_substituents=True))
factory.add_workflow_components(workflow_components.StandardConformerGenerator(max_conformers=4))
factory.dict()
oe_dataset = factory.create_dataset(dataset_name="OpenFF-benchmark-ligand-fragments-v2.0",
molecules="../2020-07-27-OpenFF-Benchmark-Ligands/sdfs/",
description="Ligand fragments generated via openff-fragmenter using openeye/ambertools for the JACS benchmark systems. These fragments are then used to fit bespoke torsion parameters for the bespokefit paper.",
tagline="Ligand fragments from the JACS benchmark systems.",
toolkit_registry=GLOBAL_TOOLKIT_REGISTRY)
oe_dataset.metadata
oe_dataset.filtered_molecules
# now put ambertools back in
GLOBAL_TOOLKIT_REGISTRY.deregister_toolkit(OpenEyeToolkitWrapper())
GLOBAL_TOOLKIT_REGISTRY.register_toolkit(AmberToolsToolkitWrapper())
print(GLOBAL_TOOLKIT_REGISTRY.registered_toolkits)
# now make the ambertools fragments
am_dataset = factory.create_dataset(dataset_name="OpenFF-benchmark-ligand-fragments-v2.0",
molecules="../2020-07-27-OpenFF-Benchmark-Ligands/sdfs/",
description="Ligand fragments generated via openff-fragmenter using openeye/ambertools for the JACS benchmark systems. These fragments are then used to fit bespoke torsion parameters for the bespokefit paper.",
tagline="Ligand fragments from the JACS benchmark systems.",
toolkit_registry=GLOBAL_TOOLKIT_REGISTRY)
am_dataset.metadata
am_dataset.filtered_molecules
# add the new datasets to deduplicate torsiondrives
new_dataset = oe_dataset + am_dataset
# load the old dataset for record deduplication
old_dataset_data = deserialize("../2020-07-27-OpenFF-Benchmark-Ligands/dataset.json.bz2")
# the filtered molecules data has changed so remove this to stop errors
del old_dataset_data["filtered_molecules"]
old_dataset = TorsiondriveDataset.parse_obj(old_dataset_data)
# add the two datasets together
combinded_dataset = new_dataset + old_dataset
# save to file
combinded_dataset.export_dataset("dataset.json.xz")
# get a list of entries which should be removed from the combined dataset and replaced with old entries
replacments = {} # (combined_id: old_entry)
for entry in old_dataset.dataset.values():
old_molecule = entry.get_off_molecule()
new_ids = combinded_dataset.get_molecule_entry(old_molecule)
if new_ids:
for id_entry in new_ids:
new_entry = combinded_dataset.dataset[id_entry]
new_molecule = new_entry.get_off_molecule()
iso, atom_map = Molecule.are_isomorphic(old_molecule, new_molecule, return_atom_map=True)
old_dihedral = entry.dihedrals[0][1:3]
new_dihedral = new_entry.dihedrals[0][1:3]
# now see if the central bond is the same
if atom_map[old_dihedral[0]] == new_dihedral[0] and atom_map[old_dihedral[1]] == new_dihedral[1] or atom_map[old_dihedral[1]] == new_dihedral[0] and atom_map[old_dihedral[0]] == new_dihedral[1]:
# log which entry should be replaced
replacments[id_entry] = entry
break
# the number of reused torsiondrives
len(replacments)
# now edit the dataset and replace the entries
for index, entry in replacments.items():
del combinded_dataset.dataset[index]
combinded_dataset.dataset[entry.index] = entry
combinded_dataset.n_molecules
combinded_dataset.n_records
for entry in replacments.values():
assert entry.index in combinded_dataset.dataset
combinded_dataset.metadata.long_description_url = "https://github.com/openforcefield/qca-dataset-submission/tree/master/submissions/2021-08-10-OpenFF-JACS-Fragments-v2.0"
combinded_dataset.metadata.submitter = "JTHorton"
# a restart cell
# combinded_dataset = TorsiondriveDataset.parse_file("dataset.json.bz2")
# reduce the max number of conformers from 10 to 4
for entry in combinded_dataset.dataset.values():
if len(entry.initial_molecules) > 4:
entry.initial_molecules = entry.initial_molecules[:4]
# collect dataset info
from openeye import oechem
import numpy as np
confs = np.array([len(mol.conformers) for mol in combinded_dataset.molecules])
print("Number of unique molecules ", combinded_dataset.n_molecules)
print("Number of filtered molecules ", combinded_dataset.n_filtered)
print("Number of torsiondrives ", combinded_dataset.n_records)
print("Number of conformers min mean max",
confs.min(), "{:6.2f}".format(confs.mean()), confs.max())
masses = []
for molecule in combinded_dataset.molecules:
oemol = molecule.to_openeye()
mass = oechem.OECalculateMolecularWeight(oemol)
masses.append(mass)
print(f'Mean molecular weight: {np.mean(np.array(masses)):.2f}')
print(f'Max molecular weight: {np.max(np.array(masses)):.2f}')
print("Charges:", sorted(set(m.total_charge/m.total_charge.unit for m in combinded_dataset.molecules)))
from pprint import pprint
pprint(combinded_dataset.metadata.dict())
for spec, obj in combinded_dataset.qc_specifications.items():
print("Spec:", spec)
pprint(obj.dict())
# export the final dataset
combinded_dataset.export_dataset("dataset.json.bz2")
combinded_dataset.molecules_to_file("dataset.smi", "smi")
combinded_dataset.visualize("dataset.pdf", columns=8)
# load the dataset to add more compute specs
dataset = load_dataset("dataset.json.bz2")
# add all xtb specs
dataset.add_qc_spec(method="gfn0xtb", basis=None, program="xtb", spec_name="gfn0xtb", spec_description="A default spec for gn0xtb")
dataset.add_qc_spec(method="gfn1xtb", basis=None, program="xtb", spec_name="gfn1xtb", spec_description="A default spec for gfn1xtb")
dataset.add_qc_spec(method="gfn2xtb", basis=None, program="xtb", spec_name="gfn2xtb", spec_description="A default spec for gfn2xtb")
dataset.add_qc_spec(method="gfnff", basis=None, program="xtb", spec_name="gfnff", spec_description="A default spec for gfnff")
# add ani2x we know that this will fail for a lot of molecules
dataset.add_qc_spec(method="ani2x", basis=None, program="torchani", spec_name="ani2x", spec_description="A default spec for ani2x")
# add all of the forcefields
dataset.add_qc_spec(method="openff-1.0.0", basis="smirnoff", spec_name="openff-1.0.0", spec_description="A default spec for openff-1.0.0", program="openmm")
dataset.add_qc_spec(method="openff-1.1.1", basis="smirnoff", spec_name="openff-1.1.1", spec_description="A default spec for openff-1.1.1", program="openmm")
dataset.add_qc_spec(method="openff-1.2.1", basis="smirnoff", spec_name="openff-1.2.1", spec_description="A default spec for openff-1.2.1", program="openmm")
dataset.add_qc_spec(method="openff-1.3.0", basis="smirnoff", spec_name="openff-1.3.0", spec_description="A default spec for openff-1.3.0", program="openmm")
dataset.add_qc_spec(method="openff-2.0.0", basis="smirnoff", spec_name="openff-2.0.0", spec_description="A default spec for openff-2.0.0", program="openmm")
dataset.add_qc_spec(method="gaff-2.11", basis="antechamber", spec_name="gaff-2.11", spec_description="A default spec for gaff-2.11", program="openmm")
for spec, obj in dataset.qc_specifications.items():
print("Spec:", spec)
pprint(obj.dict())
dataset.export_dataset("dataset.json.bz2")
```
|
github_jupyter
|
from openff.qcsubmit.factories import TorsiondriveDatasetFactory
from openff.qcsubmit import workflow_components
from openff.qcsubmit.datasets import TorsiondriveDataset, load_dataset
from openff.qcsubmit.serializers import deserialize
from openff.toolkit.topology import Molecule
from openff.toolkit.utils.toolkits import GLOBAL_TOOLKIT_REGISTRY, OpenEyeToolkitWrapper, AmberToolsToolkitWrapper
# configure the toolkit registry to only use openeye, openff-fragmenter should respect this
GLOBAL_TOOLKIT_REGISTRY.deregister_toolkit(AmberToolsToolkitWrapper())
print(GLOBAL_TOOLKIT_REGISTRY.registered_toolkits)
factory = TorsiondriveDatasetFactory()
factory.add_workflow_components(workflow_components.WBOFragmenter(keep_non_rotor_ring_substituents=True))
factory.add_workflow_components(workflow_components.StandardConformerGenerator(max_conformers=4))
factory.dict()
oe_dataset = factory.create_dataset(dataset_name="OpenFF-benchmark-ligand-fragments-v2.0",
molecules="../2020-07-27-OpenFF-Benchmark-Ligands/sdfs/",
description="Ligand fragments generated via openff-fragmenter using openeye/ambertools for the JACS benchmark systems. These fragments are then used to fit bespoke torsion parameters for the bespokefit paper.",
tagline="Ligand fragments from the JACS benchmark systems.",
toolkit_registry=GLOBAL_TOOLKIT_REGISTRY)
oe_dataset.metadata
oe_dataset.filtered_molecules
# now put ambertools back in
GLOBAL_TOOLKIT_REGISTRY.deregister_toolkit(OpenEyeToolkitWrapper())
GLOBAL_TOOLKIT_REGISTRY.register_toolkit(AmberToolsToolkitWrapper())
print(GLOBAL_TOOLKIT_REGISTRY.registered_toolkits)
# now make the ambertools fragments
am_dataset = factory.create_dataset(dataset_name="OpenFF-benchmark-ligand-fragments-v2.0",
molecules="../2020-07-27-OpenFF-Benchmark-Ligands/sdfs/",
description="Ligand fragments generated via openff-fragmenter using openeye/ambertools for the JACS benchmark systems. These fragments are then used to fit bespoke torsion parameters for the bespokefit paper.",
tagline="Ligand fragments from the JACS benchmark systems.",
toolkit_registry=GLOBAL_TOOLKIT_REGISTRY)
am_dataset.metadata
am_dataset.filtered_molecules
# add the new datasets to deduplicate torsiondrives
new_dataset = oe_dataset + am_dataset
# load the old dataset for record deduplication
old_dataset_data = deserialize("../2020-07-27-OpenFF-Benchmark-Ligands/dataset.json.bz2")
# the filtered molecules data has changed so remove this to stop errors
del old_dataset_data["filtered_molecules"]
old_dataset = TorsiondriveDataset.parse_obj(old_dataset_data)
# add the two datasets together
combinded_dataset = new_dataset + old_dataset
# save to file
combinded_dataset.export_dataset("dataset.json.xz")
# get a list of entries which should be removed from the combined dataset and replaced with old entries
replacments = {} # (combined_id: old_entry)
for entry in old_dataset.dataset.values():
old_molecule = entry.get_off_molecule()
new_ids = combinded_dataset.get_molecule_entry(old_molecule)
if new_ids:
for id_entry in new_ids:
new_entry = combinded_dataset.dataset[id_entry]
new_molecule = new_entry.get_off_molecule()
iso, atom_map = Molecule.are_isomorphic(old_molecule, new_molecule, return_atom_map=True)
old_dihedral = entry.dihedrals[0][1:3]
new_dihedral = new_entry.dihedrals[0][1:3]
# now see if the central bond is the same
if atom_map[old_dihedral[0]] == new_dihedral[0] and atom_map[old_dihedral[1]] == new_dihedral[1] or atom_map[old_dihedral[1]] == new_dihedral[0] and atom_map[old_dihedral[0]] == new_dihedral[1]:
# log which entry should be replaced
replacments[id_entry] = entry
break
# the number of reused torsiondrives
len(replacments)
# now edit the dataset and replace the entries
for index, entry in replacments.items():
del combinded_dataset.dataset[index]
combinded_dataset.dataset[entry.index] = entry
combinded_dataset.n_molecules
combinded_dataset.n_records
for entry in replacments.values():
assert entry.index in combinded_dataset.dataset
combinded_dataset.metadata.long_description_url = "https://github.com/openforcefield/qca-dataset-submission/tree/master/submissions/2021-08-10-OpenFF-JACS-Fragments-v2.0"
combinded_dataset.metadata.submitter = "JTHorton"
# a restart cell
# combinded_dataset = TorsiondriveDataset.parse_file("dataset.json.bz2")
# reduce the max number of conformers from 10 to 4
for entry in combinded_dataset.dataset.values():
if len(entry.initial_molecules) > 4:
entry.initial_molecules = entry.initial_molecules[:4]
# collect dataset info
from openeye import oechem
import numpy as np
confs = np.array([len(mol.conformers) for mol in combinded_dataset.molecules])
print("Number of unique molecules ", combinded_dataset.n_molecules)
print("Number of filtered molecules ", combinded_dataset.n_filtered)
print("Number of torsiondrives ", combinded_dataset.n_records)
print("Number of conformers min mean max",
confs.min(), "{:6.2f}".format(confs.mean()), confs.max())
masses = []
for molecule in combinded_dataset.molecules:
oemol = molecule.to_openeye()
mass = oechem.OECalculateMolecularWeight(oemol)
masses.append(mass)
print(f'Mean molecular weight: {np.mean(np.array(masses)):.2f}')
print(f'Max molecular weight: {np.max(np.array(masses)):.2f}')
print("Charges:", sorted(set(m.total_charge/m.total_charge.unit for m in combinded_dataset.molecules)))
from pprint import pprint
pprint(combinded_dataset.metadata.dict())
for spec, obj in combinded_dataset.qc_specifications.items():
print("Spec:", spec)
pprint(obj.dict())
# export the final dataset
combinded_dataset.export_dataset("dataset.json.bz2")
combinded_dataset.molecules_to_file("dataset.smi", "smi")
combinded_dataset.visualize("dataset.pdf", columns=8)
# load the dataset to add more compute specs
dataset = load_dataset("dataset.json.bz2")
# add all xtb specs
dataset.add_qc_spec(method="gfn0xtb", basis=None, program="xtb", spec_name="gfn0xtb", spec_description="A default spec for gn0xtb")
dataset.add_qc_spec(method="gfn1xtb", basis=None, program="xtb", spec_name="gfn1xtb", spec_description="A default spec for gfn1xtb")
dataset.add_qc_spec(method="gfn2xtb", basis=None, program="xtb", spec_name="gfn2xtb", spec_description="A default spec for gfn2xtb")
dataset.add_qc_spec(method="gfnff", basis=None, program="xtb", spec_name="gfnff", spec_description="A default spec for gfnff")
# add ani2x we know that this will fail for a lot of molecules
dataset.add_qc_spec(method="ani2x", basis=None, program="torchani", spec_name="ani2x", spec_description="A default spec for ani2x")
# add all of the forcefields
dataset.add_qc_spec(method="openff-1.0.0", basis="smirnoff", spec_name="openff-1.0.0", spec_description="A default spec for openff-1.0.0", program="openmm")
dataset.add_qc_spec(method="openff-1.1.1", basis="smirnoff", spec_name="openff-1.1.1", spec_description="A default spec for openff-1.1.1", program="openmm")
dataset.add_qc_spec(method="openff-1.2.1", basis="smirnoff", spec_name="openff-1.2.1", spec_description="A default spec for openff-1.2.1", program="openmm")
dataset.add_qc_spec(method="openff-1.3.0", basis="smirnoff", spec_name="openff-1.3.0", spec_description="A default spec for openff-1.3.0", program="openmm")
dataset.add_qc_spec(method="openff-2.0.0", basis="smirnoff", spec_name="openff-2.0.0", spec_description="A default spec for openff-2.0.0", program="openmm")
dataset.add_qc_spec(method="gaff-2.11", basis="antechamber", spec_name="gaff-2.11", spec_description="A default spec for gaff-2.11", program="openmm")
for spec, obj in dataset.qc_specifications.items():
print("Spec:", spec)
pprint(obj.dict())
dataset.export_dataset("dataset.json.bz2")
| 0.458106 | 0.77208 |
<a href="https://colab.research.google.com/github/anshupandey/Deep-Learning-for-structured-Data/blob/main/Copy_of_preprocessing_layers.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## The Dataset
You will use a simplified version of the PetFinder [dataset](https://www.kaggle.com/c/petfinder-adoption-prediction).
https://www.kaggle.com/c/petfinder-adoption-prediction
There are several thousand rows in the CSV. Each row describes a pet, and each column describes an attribute. You will use this information to predict if the pet will be adopted.
Following is a description of this dataset. Notice there are both numeric and categorical columns. There is a free text column which we will not use in this tutorial.
Column | Description| Feature Type | Data Type
------------|--------------------|----------------------|-----------------
Type | Type of animal (Dog, Cat) | Categorical | string
Age | Age of the pet | Numerical | integer
Breed1 | Primary breed of the pet | Categorical | string
Color1 | Color 1 of pet | Categorical | string
Color2 | Color 2 of pet | Categorical | string
MaturitySize | Size at maturity | Categorical | string
FurLength | Fur length | Categorical | string
Vaccinated | Pet has been vaccinated | Categorical | string
Sterilized | Pet has been sterilized | Categorical | string
Health | Health Condition | Categorical | string
Fee | Adoption Fee | Numerical | integer
Description | Profile write-up for this pet | Text | string
PhotoAmt | Total uploaded photos for this pet | Numerical | integer
AdoptionSpeed | Speed of adoption | Classification | integer
## Import TensorFlow and other libraries
```
import numpy as np
import pandas as pd
import tensorflow as tf
from sklearn.model_selection import train_test_split
from tensorflow.keras import layers
from tensorflow.keras.layers.experimental import preprocessing
tf.__version__
```
## Use Pandas to create a dataframe
```
import pathlib
dataset_url = 'http://storage.googleapis.com/download.tensorflow.org/data/petfinder-mini.zip'
csv_file = 'datasets/petfinder-mini/petfinder-mini.csv'
tf.keras.utils.get_file('petfinder_mini.zip', dataset_url,
extract=True, cache_dir='.')
dataframe = pd.read_csv(csv_file)
dataframe.head()
```
## Create target variable
The task in the Kaggle competition is to predict the speed at which a pet will be adopted (e.g., in the first week, the first month, the first three months, and so on). Let's simplify this for current code. Here, we will transform this into a binary classification problem, and simply predict whether the pet was adopted, or not.
After modifying the label column, 0 will indicate the pet was not adopted, and 1 will indicate it was.
```
# In the original dataset "4" indicates the pet was not adopted.
dataframe['target'] = np.where(dataframe['AdoptionSpeed']==4, 0, 1)
# Drop un-used columns.
dataframe = dataframe.drop(columns=['AdoptionSpeed', 'Description'])
```
## Split the dataframe into train, validation, and test
The dataset we downloaded was a single CSV file. we will split this into train, validation, and test sets.
```
train, test = train_test_split(dataframe, test_size=0.2)
train, val = train_test_split(train, test_size=0.2)
print(len(train), 'train examples')
print(len(val), 'validation examples')
print(len(test), 'test examples')
```
## Create an input pipeline using tf.data
Next, we will wrap the dataframes with [tf.data](https://www.tensorflow.org/guide/datasets), in order to shuffle and batch the data. If we were working with a very large CSV file (so large that it does not fit into memory), we would use tf.data to read it from disk directly.
```
# A utility method to create a tf.data dataset from a Pandas Dataframe
def df_to_dataset(dataframe, shuffle=True, batch_size=32):
dataframe = dataframe.copy()
labels = dataframe.pop('target')
ds = tf.data.Dataset.from_tensor_slices((dict(dataframe), labels))
if shuffle:
ds = ds.shuffle(buffer_size=len(dataframe))
ds = ds.batch(batch_size)
ds = ds.prefetch(batch_size)
return ds
```
Now that you have created the input pipeline, let's call it to see the format of the data it returns. You have used a small batch size to keep the output readable.
```
batch_size = 5
train_ds = df_to_dataset(train, batch_size=batch_size)
[(train_features, label_batch)] = train_ds.take(1)
print('Every feature:', list(train_features.keys()))
print('A batch of ages:', train_features['Age'])
print('A batch of targets:', label_batch )
```
You can see that the dataset returns a dictionary of column names (from the dataframe) that map to column values from rows in the dataframe.
## Demonstrate the use of preprocessing layers.
The Keras preprocessing layers API allows you to build Keras-native input processing pipelines. You will use 3 preprocessing layers to demonstrate the feature preprocessing code.
* [`Normalization`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/experimental/preprocessing/Normalization) - Feature-wise normalization of the data.
* [`CategoryEncoding`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/experimental/preprocessing/CategoryEncoding) - Category encoding layer.
* [`StringLookup`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/experimental/preprocessing/StringLookup) - Maps strings from a vocabulary to integer indices.
* [`IntegerLookup`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/experimental/preprocessing/IntegerLookup) - Maps integers from a vocabulary to integer indices.
You can find a list of available preprocessing layers [here](https://www.tensorflow.org/api_docs/python/tf/keras/layers/experimental/preprocessing).
https://www.tensorflow.org/api_docs/python/tf/keras/layers/experimental/preprocessing
### Numeric columns
For each of the Numeric feature, you will use a Normalization() layer to make sure the mean of each feature is 0 and its standard deviation is 1.
`get_normalization_layer` function returns a layer which applies featurewise normalization to numerical features.
```
def get_normalization_layer(name, dataset):
# Create a Normalization layer for our feature.
normalizer = preprocessing.Normalization(axis=None)
# Prepare a Dataset that only yields our feature.
feature_ds = dataset.map(lambda x, y: x[name])
# Learn the statistics of the data.
normalizer.adapt(feature_ds)
return normalizer
photo_count_col = train_features['PhotoAmt']
layer = get_normalization_layer('PhotoAmt', train_ds)
layer(photo_count_col)
```
Note: If you many numeric features (hundreds, or more), it is more efficient to concatenate them first and use a single [normalization](https://www.tensorflow.org/api_docs/python/tf/keras/layers/experimental/preprocessing/Normalization) layer.
### Categorical columns
In this dataset, Type is represented as a string (e.g. 'Dog', or 'Cat'). You cannot feed strings directly to a model. The preprocessing layer takes care of representing strings as a one-hot vector.
`get_category_encoding_layer` function returns a layer which maps values from a vocabulary to integer indices and one-hot encodes the features.
```
def get_category_encoding_layer(name, dataset, dtype, max_tokens=None):
# Create a StringLookup layer which will turn strings into integer indices
if dtype == 'string':
index = preprocessing.StringLookup(max_tokens=max_tokens)
else:
index = preprocessing.IntegerLookup(max_tokens=max_tokens)
# Prepare a Dataset that only yields our feature
feature_ds = dataset.map(lambda x, y: x[name])
# Learn the set of possible values and assign them a fixed integer index.
index.adapt(feature_ds)
# Create a Discretization for our integer indices.
encoder = preprocessing.CategoryEncoding(num_tokens=index.vocabulary_size())
# Apply one-hot encoding to our indices. The lambda function captures the
# layer so we can use them, or include them in the functional model later.
return lambda feature: encoder(index(feature))
type_col = train_features['Type']
layer = get_category_encoding_layer('Type', train_ds, 'string')
layer(type_col)
```
Often, you don't want to feed a number directly into the model, but instead use a one-hot encoding of those inputs. Consider raw data that represents a pet's age.
```
type_col = train_features['Age']
category_encoding_layer = get_category_encoding_layer('Age', train_ds,
'int64', 5)
category_encoding_layer(type_col)
```
## Choose which columns to use
You have seen how to use several types of preprocessing layers. Now you will use them to train a model. You will be using [Keras-functional API](https://www.tensorflow.org/guide/keras/functional) to build the model. The Keras functional API is a way to create models that are more flexible than the [tf.keras.Sequential](https://www.tensorflow.org/api_docs/python/tf/keras/Sequential) API.
The goal of this tutorial is to show you the complete code (e.g. mechanics) needed to work with preprocessing layers. A few columns have been selected arbitrarily to train our model.
Key point: If your aim is to build an accurate model, try a larger dataset of your own, and think carefully about which features are the most meaningful to include, and how they should be represented.
Earlier, you used a small batch size to demonstrate the input pipeline. Let's now create a new input pipeline with a larger batch size.
```
batch_size = 256
train_ds = df_to_dataset(train, batch_size=batch_size)
val_ds = df_to_dataset(val, shuffle=False, batch_size=batch_size)
test_ds = df_to_dataset(test, shuffle=False, batch_size=batch_size)
all_inputs = []
encoded_features = []
# Numeric features.
for header in ['PhotoAmt', 'Fee']:
numeric_col = tf.keras.Input(shape=(1,), name=header)
normalization_layer = get_normalization_layer(header, train_ds)
encoded_numeric_col = normalization_layer(numeric_col)
all_inputs.append(numeric_col)
encoded_features.append(encoded_numeric_col)
# Categorical features encoded as integers.
age_col = tf.keras.Input(shape=(1,), name='Age', dtype='int64')
encoding_layer = get_category_encoding_layer('Age', train_ds, dtype='int64',
max_tokens=5)
encoded_age_col = encoding_layer(age_col)
all_inputs.append(age_col)
encoded_features.append(encoded_age_col)
# Categorical features encoded as string.
categorical_cols = ['Type', 'Color1', 'Color2', 'Gender', 'MaturitySize',
'FurLength', 'Vaccinated', 'Sterilized', 'Health', 'Breed1']
for header in categorical_cols:
categorical_col = tf.keras.Input(shape=(1,), name=header, dtype='string')
encoding_layer = get_category_encoding_layer(header, train_ds, dtype='string',
max_tokens=5)
encoded_categorical_col = encoding_layer(categorical_col)
all_inputs.append(categorical_col)
encoded_features.append(encoded_categorical_col)
```
## Create, compile, and train the model
Now you can create our end-to-end model.
```
all_features = tf.keras.layers.concatenate(encoded_features)
x = tf.keras.layers.Dense(32, activation="relu")(all_features)
x = tf.keras.layers.Dropout(0.5)(x)
output = tf.keras.layers.Dense(1)(x)
model = tf.keras.Model(all_inputs, output)
model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=["accuracy"])
```
Let's visualize our connectivity graph:
```
# rankdir='LR' is used to make the graph horizontal.
tf.keras.utils.plot_model(model, show_shapes=True, rankdir="LR")
```
### Train the model
```
model.fit(train_ds, epochs=10, validation_data=val_ds)
loss, accuracy = model.evaluate(test_ds)
print("Accuracy", accuracy)
```
## Inference on new data
Key point: The model you have developed can now classify a row from a CSV file directly, because the preprocessing code is included inside the model itself.
You can now save and reload the Keras model. Follow the tutorial [here](https://www.tensorflow.org/tutorials/keras/save_and_load) for more information on TensorFlow models.
```
model.save('my_pet_classifier')
reloaded_model = tf.keras.models.load_model('my_pet_classifier')
```
To get a prediction for a new sample, you can simply call `model.predict()`. There are just two things you need to do:
1. Wrap scalars into a list so as to have a batch dimension (models only process batches of data, not single samples)
2. Call `convert_to_tensor` on each feature
```
sample = {
'Type': 'Cat',
'Age': 3,
'Breed1': 'Tabby',
'Gender': 'Male',
'Color1': 'Black',
'Color2': 'White',
'MaturitySize': 'Small',
'FurLength': 'Short',
'Vaccinated': 'No',
'Sterilized': 'No',
'Health': 'Healthy',
'Fee': 100,
'PhotoAmt': 2,
}
input_dict = {name: tf.convert_to_tensor([value]) for name, value in sample.items()}
predictions = reloaded_model.predict(input_dict)
prob = tf.nn.sigmoid(predictions[0])
print(
"This particular pet had a %.1f percent probability "
"of getting adopted." % (100 * prob)
)
```
Key point: You will typically see best results with deep learning with larger and more complex datasets. When working with a small dataset like this one, we recommend using a decision tree or random forest as a strong baseline. The goal of this tutorial is to demonstrate the mechanics of working with structured data, so you have code to use as a starting point when working with your own datasets in the future.
## Next steps
The best way to learn more about classifying structured data is to try it yourself. You may want to find another dataset to work with, and training a model to classify it using code similar to the above. To improve accuracy, think carefully about which features to include in your model, and how they should be represented.
|
github_jupyter
|
import numpy as np
import pandas as pd
import tensorflow as tf
from sklearn.model_selection import train_test_split
from tensorflow.keras import layers
from tensorflow.keras.layers.experimental import preprocessing
tf.__version__
import pathlib
dataset_url = 'http://storage.googleapis.com/download.tensorflow.org/data/petfinder-mini.zip'
csv_file = 'datasets/petfinder-mini/petfinder-mini.csv'
tf.keras.utils.get_file('petfinder_mini.zip', dataset_url,
extract=True, cache_dir='.')
dataframe = pd.read_csv(csv_file)
dataframe.head()
# In the original dataset "4" indicates the pet was not adopted.
dataframe['target'] = np.where(dataframe['AdoptionSpeed']==4, 0, 1)
# Drop un-used columns.
dataframe = dataframe.drop(columns=['AdoptionSpeed', 'Description'])
train, test = train_test_split(dataframe, test_size=0.2)
train, val = train_test_split(train, test_size=0.2)
print(len(train), 'train examples')
print(len(val), 'validation examples')
print(len(test), 'test examples')
# A utility method to create a tf.data dataset from a Pandas Dataframe
def df_to_dataset(dataframe, shuffle=True, batch_size=32):
dataframe = dataframe.copy()
labels = dataframe.pop('target')
ds = tf.data.Dataset.from_tensor_slices((dict(dataframe), labels))
if shuffle:
ds = ds.shuffle(buffer_size=len(dataframe))
ds = ds.batch(batch_size)
ds = ds.prefetch(batch_size)
return ds
batch_size = 5
train_ds = df_to_dataset(train, batch_size=batch_size)
[(train_features, label_batch)] = train_ds.take(1)
print('Every feature:', list(train_features.keys()))
print('A batch of ages:', train_features['Age'])
print('A batch of targets:', label_batch )
def get_normalization_layer(name, dataset):
# Create a Normalization layer for our feature.
normalizer = preprocessing.Normalization(axis=None)
# Prepare a Dataset that only yields our feature.
feature_ds = dataset.map(lambda x, y: x[name])
# Learn the statistics of the data.
normalizer.adapt(feature_ds)
return normalizer
photo_count_col = train_features['PhotoAmt']
layer = get_normalization_layer('PhotoAmt', train_ds)
layer(photo_count_col)
def get_category_encoding_layer(name, dataset, dtype, max_tokens=None):
# Create a StringLookup layer which will turn strings into integer indices
if dtype == 'string':
index = preprocessing.StringLookup(max_tokens=max_tokens)
else:
index = preprocessing.IntegerLookup(max_tokens=max_tokens)
# Prepare a Dataset that only yields our feature
feature_ds = dataset.map(lambda x, y: x[name])
# Learn the set of possible values and assign them a fixed integer index.
index.adapt(feature_ds)
# Create a Discretization for our integer indices.
encoder = preprocessing.CategoryEncoding(num_tokens=index.vocabulary_size())
# Apply one-hot encoding to our indices. The lambda function captures the
# layer so we can use them, or include them in the functional model later.
return lambda feature: encoder(index(feature))
type_col = train_features['Type']
layer = get_category_encoding_layer('Type', train_ds, 'string')
layer(type_col)
type_col = train_features['Age']
category_encoding_layer = get_category_encoding_layer('Age', train_ds,
'int64', 5)
category_encoding_layer(type_col)
batch_size = 256
train_ds = df_to_dataset(train, batch_size=batch_size)
val_ds = df_to_dataset(val, shuffle=False, batch_size=batch_size)
test_ds = df_to_dataset(test, shuffle=False, batch_size=batch_size)
all_inputs = []
encoded_features = []
# Numeric features.
for header in ['PhotoAmt', 'Fee']:
numeric_col = tf.keras.Input(shape=(1,), name=header)
normalization_layer = get_normalization_layer(header, train_ds)
encoded_numeric_col = normalization_layer(numeric_col)
all_inputs.append(numeric_col)
encoded_features.append(encoded_numeric_col)
# Categorical features encoded as integers.
age_col = tf.keras.Input(shape=(1,), name='Age', dtype='int64')
encoding_layer = get_category_encoding_layer('Age', train_ds, dtype='int64',
max_tokens=5)
encoded_age_col = encoding_layer(age_col)
all_inputs.append(age_col)
encoded_features.append(encoded_age_col)
# Categorical features encoded as string.
categorical_cols = ['Type', 'Color1', 'Color2', 'Gender', 'MaturitySize',
'FurLength', 'Vaccinated', 'Sterilized', 'Health', 'Breed1']
for header in categorical_cols:
categorical_col = tf.keras.Input(shape=(1,), name=header, dtype='string')
encoding_layer = get_category_encoding_layer(header, train_ds, dtype='string',
max_tokens=5)
encoded_categorical_col = encoding_layer(categorical_col)
all_inputs.append(categorical_col)
encoded_features.append(encoded_categorical_col)
all_features = tf.keras.layers.concatenate(encoded_features)
x = tf.keras.layers.Dense(32, activation="relu")(all_features)
x = tf.keras.layers.Dropout(0.5)(x)
output = tf.keras.layers.Dense(1)(x)
model = tf.keras.Model(all_inputs, output)
model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=["accuracy"])
# rankdir='LR' is used to make the graph horizontal.
tf.keras.utils.plot_model(model, show_shapes=True, rankdir="LR")
model.fit(train_ds, epochs=10, validation_data=val_ds)
loss, accuracy = model.evaluate(test_ds)
print("Accuracy", accuracy)
model.save('my_pet_classifier')
reloaded_model = tf.keras.models.load_model('my_pet_classifier')
sample = {
'Type': 'Cat',
'Age': 3,
'Breed1': 'Tabby',
'Gender': 'Male',
'Color1': 'Black',
'Color2': 'White',
'MaturitySize': 'Small',
'FurLength': 'Short',
'Vaccinated': 'No',
'Sterilized': 'No',
'Health': 'Healthy',
'Fee': 100,
'PhotoAmt': 2,
}
input_dict = {name: tf.convert_to_tensor([value]) for name, value in sample.items()}
predictions = reloaded_model.predict(input_dict)
prob = tf.nn.sigmoid(predictions[0])
print(
"This particular pet had a %.1f percent probability "
"of getting adopted." % (100 * prob)
)
| 0.793866 | 0.985257 |
```
import numpy as np
from matplotlib import pyplot as plt
from sklearn import preprocessing
import wfdb
import copy as cp
import scipy.signal as signal
import pickle
from sklearn import preprocessing
from tqdm import tqdm
import os
import re
import pandas as pd
import csv
from sklearn.linear_model import LogisticRegression
from sklearn import neighbors
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.svm import SVC
from sklearn.ensemble import StackingClassifier
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.model_selection import KFold
from sklearn.metrics import accuracy_score
from sklearn.model_selection import cross_val_score
from time import time
import timeit
record_list = [] # Initialize the array that will hold the list of our records
records = 'mit-bih-dataframes/subject_list.csv' # Get our record list like we did in the initial extraction
with open(records) as rfile:# Load our records into the array
for record in rfile:
record = record[0:-1] # The -1 removes the newline ("\n") character from the string
record_list.append(record)
dfdic = {}
for idx, x in enumerate(record_list):
dfdic[x] = pd.read_csv('mit-bih-features/'+x+ '.csv', index_col=0)
subject_df = pd.DataFrame()
for idx, x in enumerate(record_list):
subject_df = pd.concat([subject_df, dfdic[x]])
subject_df = subject_df.drop(["Unnamed: 0.1"], axis=1)
subject_df['Mappedrhythmlabels'] = subject_df['rhythmLabel'].map({'Normal':0, 'Other':0, 'AFIB':1})
subject_df.head()
models_dict = {
'Logistic Regression': LogisticRegression(solver='liblinear'),
'LDA': LinearDiscriminantAnalysis(),
'QDA': QuadraticDiscriminantAnalysis(),
'KNN-CV': neighbors.KNeighborsClassifier(n_neighbors=8),
'Decision Tree': DecisionTreeClassifier(max_depth=10),
'Bagging': RandomForestClassifier(max_features=15, random_state=3),
'Random Forest': RandomForestClassifier(max_features=4, random_state=3),
'Adaptive Boosting': AdaBoostClassifier(n_estimators=500, learning_rate=0.8, random_state=3),
'Gradient Boosting': GradientBoostingClassifier(n_estimators=500, learning_rate=0.1 , max_depth=8, random_state=3),
'SVCLinear': SVC(C=1, kernel='linear', random_state=3),
'SVCRadial': SVC(C=5, gamma=0.001 , kernel='rbf')
}
base_models = [('Logistic Regression', models_dict['Logistic Regression']), ('LDA', models_dict['LDA']), ('QDA', models_dict['QDA']), ('KNN-CV', models_dict['KNN-CV']),
('Decision Tree', models_dict['Decision Tree']), ('Bagging', models_dict['Bagging']), ('Random Forest', models_dict['Random Forest']), ('Adaptive Boosting', models_dict['Adaptive Boosting']),
('Gradient Boosting', models_dict['Gradient Boosting']), ('SVCLinear', models_dict['SVCLinear']), ('SVCRadial', models_dict['SVCRadial'])]
meta_model = LogisticRegression(solver='liblinear')
stacking_model = StackingClassifier(estimators=base_models, final_estimator=meta_model, passthrough=True, cv=5, verbose=2)
kf = KFold(n_splits=5, random_state=3, shuffle=True)
model = stacking_model
start_time = timeit.default_timer()
acc_score = []
Truth = []
Output = []
for train_index, test_index in kf.split(subject_df):
X_train = subject_df.iloc[train_index, 2:17]
X_test = subject_df.iloc[test_index, 2:17]
Y_train = subject_df.iloc[train_index, -1]
Y_test = subject_df.iloc[test_index, -1]
model.fit(X_train, Y_train)
pred_values = model.predict(X_test)
acc = accuracy_score(Y_test, pred_values)
acc_score.append(acc)
Truth.extend(Y_test.values.reshape(Y_test.shape[0]))
Output.extend(pred_values)
elapsed = timeit.default_timer() - start_time
print("---Run time is %s seconds ---" % elapsed)
print()
print('Accuracy of each fold: \n {}'.format(acc_score))
print("Avg accuracy: {}".format(np.mean(acc_score)))
print('Std of accuracy : \n{}'.format(np.std(acc_score)))
print()
print(confusion_matrix(Truth, Output))
print()
print(classification_report(Truth, Output))
cm = confusion_matrix(Truth, Output)
sensitivity = cm[0][0]/(cm[0][0]+cm[0][1])
specificity = cm[1][1]/(cm[1][0]+cm[1][1])
precision = (cm[0][0])/(cm[0][0]+cm[1][0])
f1_score = (2*precision*sensitivity)/(precision+sensitivity)
print(sensitivity)
print(specificity)
print(precision)
print(f1_score)
base_models = [('Logistic Regression', models_dict['Logistic Regression']), ('KNN-CV', models_dict['KNN-CV']),
('Decision Tree', models_dict['Decision Tree']), ('Bagging', models_dict['Bagging']), ('Random Forest', models_dict['Random Forest']), ('Adaptive Boosting', models_dict['Adaptive Boosting']),
('Gradient Boosting', models_dict['Gradient Boosting']), ('SVCLinear', models_dict['SVCLinear'])]
meta_model = LogisticRegression(solver='liblinear')
stacking_model = StackingClassifier(estimators=base_models, final_estimator=meta_model, passthrough=True, cv=5, verbose=2)
kf = KFold(n_splits=5, random_state=3, shuffle=True)
model = stacking_model
start_time = timeit.default_timer()
acc_score = []
Truth = []
Output = []
for train_index, test_index in kf.split(subject_df):
X_train = subject_df.iloc[train_index, 2:17]
X_test = subject_df.iloc[test_index, 2:17]
Y_train = subject_df.iloc[train_index, -1]
Y_test = subject_df.iloc[test_index, -1]
model.fit(X_train, Y_train)
pred_values = model.predict(X_test)
acc = accuracy_score(Y_test, pred_values)
acc_score.append(acc)
Truth.extend(Y_test.values.reshape(Y_test.shape[0]))
Output.extend(pred_values)
elapsed = timeit.default_timer() - start_time
print("---Run time is %s seconds ---" % elapsed)
print()
print('Accuracy of each fold: \n {}'.format(acc_score))
print("Avg accuracy: {}".format(np.mean(acc_score)))
print('Std of accuracy : \n{}'.format(np.std(acc_score)))
print()
print(confusion_matrix(Truth, Output))
print()
print(classification_report(Truth, Output))
cm = confusion_matrix(Truth, Output)
sensitivity = cm[0][0]/(cm[0][0]+cm[0][1])
specificity = cm[1][1]/(cm[1][0]+cm[1][1])
precision = (cm[0][0])/(cm[0][0]+cm[1][0])
f1_score = (2*precision*sensitivity)/(precision+sensitivity)
print(sensitivity)
print(specificity)
print(precision)
print(f1_score)
base_models = []
meta_model = LogisticRegression(solver='liblinear')
stacking_model = StackingClassifier(estimators=base_models, final_estimator=meta_model, passthrough=True, cv=5, verbose=2)
kf = KFold(n_splits=5, random_state=3, shuffle=True)
model = stacking_model
start_time = timeit.default_timer()
acc_score = []
Truth = []
Output = []
for train_index, test_index in kf.split(subject_df):
X_train = subject_df.iloc[train_index, 2:17]
X_test = subject_df.iloc[test_index, 2:17]
Y_train = subject_df.iloc[train_index, -1]
Y_test = subject_df.iloc[test_index, -1]
model.fit(X_train, Y_train)
pred_values = model.predict(X_test)
acc = accuracy_score(Y_test, pred_values)
acc_score.append(acc)
Truth.extend(Y_test.values.reshape(Y_test.shape[0]))
Output.extend(pred_values)
elapsed = timeit.default_timer() - start_time
print("---Run time is %s seconds ---" % elapsed)
print()
print('Accuracy of each fold: \n {}'.format(acc_score))
print("Avg accuracy: {}".format(np.mean(acc_score)))
print('Std of accuracy : \n{}'.format(np.std(acc_score)))
print()
print(confusion_matrix(Truth, Output))
print()
print(classification_report(Truth, Output))
cm = confusion_matrix(Truth, Output)
sensitivity = cm[0][0]/(cm[0][0]+cm[0][1])
specificity = cm[1][1]/(cm[1][0]+cm[1][1])
precision = (cm[0][0])/(cm[0][0]+cm[1][0])
f1_score = (2*precision*sensitivity)/(precision+sensitivity)
print(sensitivity)
print(specificity)
print(precision)
print(f1_score)
base_models = [('Logistic Regression', models_dict['Logistic Regression']), ('LDA', models_dict['LDA']), ('QDA', models_dict['QDA']), ('KNN-CV', models_dict['KNN-CV']), ('Decision Tree', models_dict['Decision Tree']) ]
meta_model = LogisticRegression(solver='liblinear')
stacking_model = StackingClassifier(estimators=base_models, final_estimator=meta_model, passthrough=True, cv=5, verbose=2)
kf = KFold(n_splits=5, random_state=3, shuffle=True)
model = stacking_model
start_time = timeit.default_timer()
acc_score = []
Truth = []
Output = []
for train_index, test_index in kf.split(subject_df):
X_train = subject_df.iloc[train_index, 2:17]
X_test = subject_df.iloc[test_index, 2:17]
Y_train = subject_df.iloc[train_index, -1]
Y_test = subject_df.iloc[test_index, -1]
model.fit(X_train, Y_train)
pred_values = model.predict(X_test)
acc = accuracy_score(Y_test, pred_values)
acc_score.append(acc)
Truth.extend(Y_test.values.reshape(Y_test.shape[0]))
Output.extend(pred_values)
elapsed = timeit.default_timer() - start_time
print("---Run time is %s seconds ---" % elapsed)
print()
print('Accuracy of each fold: \n {}'.format(acc_score))
print("Avg accuracy: {}".format(np.mean(acc_score)))
print('Std of accuracy : \n{}'.format(np.std(acc_score)))
print()
print(confusion_matrix(Truth, Output))
print()
print(classification_report(Truth, Output))
cm = confusion_matrix(Truth, Output)
sensitivity = cm[0][0]/(cm[0][0]+cm[0][1])
specificity = cm[1][1]/(cm[1][0]+cm[1][1])
precision = (cm[0][0])/(cm[0][0]+cm[1][0])
f1_score = (2*precision*sensitivity)/(precision+sensitivity)
print(sensitivity)
print(specificity)
print(precision)
print(f1_score)
```
|
github_jupyter
|
import numpy as np
from matplotlib import pyplot as plt
from sklearn import preprocessing
import wfdb
import copy as cp
import scipy.signal as signal
import pickle
from sklearn import preprocessing
from tqdm import tqdm
import os
import re
import pandas as pd
import csv
from sklearn.linear_model import LogisticRegression
from sklearn import neighbors
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.svm import SVC
from sklearn.ensemble import StackingClassifier
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.model_selection import KFold
from sklearn.metrics import accuracy_score
from sklearn.model_selection import cross_val_score
from time import time
import timeit
record_list = [] # Initialize the array that will hold the list of our records
records = 'mit-bih-dataframes/subject_list.csv' # Get our record list like we did in the initial extraction
with open(records) as rfile:# Load our records into the array
for record in rfile:
record = record[0:-1] # The -1 removes the newline ("\n") character from the string
record_list.append(record)
dfdic = {}
for idx, x in enumerate(record_list):
dfdic[x] = pd.read_csv('mit-bih-features/'+x+ '.csv', index_col=0)
subject_df = pd.DataFrame()
for idx, x in enumerate(record_list):
subject_df = pd.concat([subject_df, dfdic[x]])
subject_df = subject_df.drop(["Unnamed: 0.1"], axis=1)
subject_df['Mappedrhythmlabels'] = subject_df['rhythmLabel'].map({'Normal':0, 'Other':0, 'AFIB':1})
subject_df.head()
models_dict = {
'Logistic Regression': LogisticRegression(solver='liblinear'),
'LDA': LinearDiscriminantAnalysis(),
'QDA': QuadraticDiscriminantAnalysis(),
'KNN-CV': neighbors.KNeighborsClassifier(n_neighbors=8),
'Decision Tree': DecisionTreeClassifier(max_depth=10),
'Bagging': RandomForestClassifier(max_features=15, random_state=3),
'Random Forest': RandomForestClassifier(max_features=4, random_state=3),
'Adaptive Boosting': AdaBoostClassifier(n_estimators=500, learning_rate=0.8, random_state=3),
'Gradient Boosting': GradientBoostingClassifier(n_estimators=500, learning_rate=0.1 , max_depth=8, random_state=3),
'SVCLinear': SVC(C=1, kernel='linear', random_state=3),
'SVCRadial': SVC(C=5, gamma=0.001 , kernel='rbf')
}
base_models = [('Logistic Regression', models_dict['Logistic Regression']), ('LDA', models_dict['LDA']), ('QDA', models_dict['QDA']), ('KNN-CV', models_dict['KNN-CV']),
('Decision Tree', models_dict['Decision Tree']), ('Bagging', models_dict['Bagging']), ('Random Forest', models_dict['Random Forest']), ('Adaptive Boosting', models_dict['Adaptive Boosting']),
('Gradient Boosting', models_dict['Gradient Boosting']), ('SVCLinear', models_dict['SVCLinear']), ('SVCRadial', models_dict['SVCRadial'])]
meta_model = LogisticRegression(solver='liblinear')
stacking_model = StackingClassifier(estimators=base_models, final_estimator=meta_model, passthrough=True, cv=5, verbose=2)
kf = KFold(n_splits=5, random_state=3, shuffle=True)
model = stacking_model
start_time = timeit.default_timer()
acc_score = []
Truth = []
Output = []
for train_index, test_index in kf.split(subject_df):
X_train = subject_df.iloc[train_index, 2:17]
X_test = subject_df.iloc[test_index, 2:17]
Y_train = subject_df.iloc[train_index, -1]
Y_test = subject_df.iloc[test_index, -1]
model.fit(X_train, Y_train)
pred_values = model.predict(X_test)
acc = accuracy_score(Y_test, pred_values)
acc_score.append(acc)
Truth.extend(Y_test.values.reshape(Y_test.shape[0]))
Output.extend(pred_values)
elapsed = timeit.default_timer() - start_time
print("---Run time is %s seconds ---" % elapsed)
print()
print('Accuracy of each fold: \n {}'.format(acc_score))
print("Avg accuracy: {}".format(np.mean(acc_score)))
print('Std of accuracy : \n{}'.format(np.std(acc_score)))
print()
print(confusion_matrix(Truth, Output))
print()
print(classification_report(Truth, Output))
cm = confusion_matrix(Truth, Output)
sensitivity = cm[0][0]/(cm[0][0]+cm[0][1])
specificity = cm[1][1]/(cm[1][0]+cm[1][1])
precision = (cm[0][0])/(cm[0][0]+cm[1][0])
f1_score = (2*precision*sensitivity)/(precision+sensitivity)
print(sensitivity)
print(specificity)
print(precision)
print(f1_score)
base_models = [('Logistic Regression', models_dict['Logistic Regression']), ('KNN-CV', models_dict['KNN-CV']),
('Decision Tree', models_dict['Decision Tree']), ('Bagging', models_dict['Bagging']), ('Random Forest', models_dict['Random Forest']), ('Adaptive Boosting', models_dict['Adaptive Boosting']),
('Gradient Boosting', models_dict['Gradient Boosting']), ('SVCLinear', models_dict['SVCLinear'])]
meta_model = LogisticRegression(solver='liblinear')
stacking_model = StackingClassifier(estimators=base_models, final_estimator=meta_model, passthrough=True, cv=5, verbose=2)
kf = KFold(n_splits=5, random_state=3, shuffle=True)
model = stacking_model
start_time = timeit.default_timer()
acc_score = []
Truth = []
Output = []
for train_index, test_index in kf.split(subject_df):
X_train = subject_df.iloc[train_index, 2:17]
X_test = subject_df.iloc[test_index, 2:17]
Y_train = subject_df.iloc[train_index, -1]
Y_test = subject_df.iloc[test_index, -1]
model.fit(X_train, Y_train)
pred_values = model.predict(X_test)
acc = accuracy_score(Y_test, pred_values)
acc_score.append(acc)
Truth.extend(Y_test.values.reshape(Y_test.shape[0]))
Output.extend(pred_values)
elapsed = timeit.default_timer() - start_time
print("---Run time is %s seconds ---" % elapsed)
print()
print('Accuracy of each fold: \n {}'.format(acc_score))
print("Avg accuracy: {}".format(np.mean(acc_score)))
print('Std of accuracy : \n{}'.format(np.std(acc_score)))
print()
print(confusion_matrix(Truth, Output))
print()
print(classification_report(Truth, Output))
cm = confusion_matrix(Truth, Output)
sensitivity = cm[0][0]/(cm[0][0]+cm[0][1])
specificity = cm[1][1]/(cm[1][0]+cm[1][1])
precision = (cm[0][0])/(cm[0][0]+cm[1][0])
f1_score = (2*precision*sensitivity)/(precision+sensitivity)
print(sensitivity)
print(specificity)
print(precision)
print(f1_score)
base_models = []
meta_model = LogisticRegression(solver='liblinear')
stacking_model = StackingClassifier(estimators=base_models, final_estimator=meta_model, passthrough=True, cv=5, verbose=2)
kf = KFold(n_splits=5, random_state=3, shuffle=True)
model = stacking_model
start_time = timeit.default_timer()
acc_score = []
Truth = []
Output = []
for train_index, test_index in kf.split(subject_df):
X_train = subject_df.iloc[train_index, 2:17]
X_test = subject_df.iloc[test_index, 2:17]
Y_train = subject_df.iloc[train_index, -1]
Y_test = subject_df.iloc[test_index, -1]
model.fit(X_train, Y_train)
pred_values = model.predict(X_test)
acc = accuracy_score(Y_test, pred_values)
acc_score.append(acc)
Truth.extend(Y_test.values.reshape(Y_test.shape[0]))
Output.extend(pred_values)
elapsed = timeit.default_timer() - start_time
print("---Run time is %s seconds ---" % elapsed)
print()
print('Accuracy of each fold: \n {}'.format(acc_score))
print("Avg accuracy: {}".format(np.mean(acc_score)))
print('Std of accuracy : \n{}'.format(np.std(acc_score)))
print()
print(confusion_matrix(Truth, Output))
print()
print(classification_report(Truth, Output))
cm = confusion_matrix(Truth, Output)
sensitivity = cm[0][0]/(cm[0][0]+cm[0][1])
specificity = cm[1][1]/(cm[1][0]+cm[1][1])
precision = (cm[0][0])/(cm[0][0]+cm[1][0])
f1_score = (2*precision*sensitivity)/(precision+sensitivity)
print(sensitivity)
print(specificity)
print(precision)
print(f1_score)
base_models = [('Logistic Regression', models_dict['Logistic Regression']), ('LDA', models_dict['LDA']), ('QDA', models_dict['QDA']), ('KNN-CV', models_dict['KNN-CV']), ('Decision Tree', models_dict['Decision Tree']) ]
meta_model = LogisticRegression(solver='liblinear')
stacking_model = StackingClassifier(estimators=base_models, final_estimator=meta_model, passthrough=True, cv=5, verbose=2)
kf = KFold(n_splits=5, random_state=3, shuffle=True)
model = stacking_model
start_time = timeit.default_timer()
acc_score = []
Truth = []
Output = []
for train_index, test_index in kf.split(subject_df):
X_train = subject_df.iloc[train_index, 2:17]
X_test = subject_df.iloc[test_index, 2:17]
Y_train = subject_df.iloc[train_index, -1]
Y_test = subject_df.iloc[test_index, -1]
model.fit(X_train, Y_train)
pred_values = model.predict(X_test)
acc = accuracy_score(Y_test, pred_values)
acc_score.append(acc)
Truth.extend(Y_test.values.reshape(Y_test.shape[0]))
Output.extend(pred_values)
elapsed = timeit.default_timer() - start_time
print("---Run time is %s seconds ---" % elapsed)
print()
print('Accuracy of each fold: \n {}'.format(acc_score))
print("Avg accuracy: {}".format(np.mean(acc_score)))
print('Std of accuracy : \n{}'.format(np.std(acc_score)))
print()
print(confusion_matrix(Truth, Output))
print()
print(classification_report(Truth, Output))
cm = confusion_matrix(Truth, Output)
sensitivity = cm[0][0]/(cm[0][0]+cm[0][1])
specificity = cm[1][1]/(cm[1][0]+cm[1][1])
precision = (cm[0][0])/(cm[0][0]+cm[1][0])
f1_score = (2*precision*sensitivity)/(precision+sensitivity)
print(sensitivity)
print(specificity)
print(precision)
print(f1_score)
| 0.48438 | 0.385172 |
# DECISION TREE CLASSIFIER ##
For the given ‘Iris’ dataset, create the Decision Tree classifier and
visualize it graphically. The purpose is if we feed any new data to this
classifier, it would be able to predict the right class accordingly.
```
# Importing certain libraries
import pandas as pd
import numpy as np
import seaborn as sb
import matplotlib.pyplot as plt
%matplotlib inline
import sklearn.datasets as datasets
```
## Understanding the data ##
```
# Importing the dataset
data = pd.read_csv(r"D:\TSF\Task 4\Iris.csv")
# Displaying the dataset
data.head(151)
# For calculating the ideal number of clusters, we will unique functions to classify the number of species present
data["Species"].unique()
# Gathering the basic information
data.info()
# Data types
data.dtypes
# Checking for any null values or missing values
data.isnull().any()
```
Since, there are no null or missing values present, therefore we can move further for data exploration
## Data Visualization ##
```
# First, using seaborn pairplot for data visualisation
sb.set(style = "whitegrid")
plt.figure(figsize = (20, 10))
sb.pairplot(data, hue = "Species")
# Second, using seaborn heatmaps for data visualisation
sb.heatmap(data.corr(), annot = True, fmt = ".3g", linewidth = 0.5, linecolor = "Black", cmap = "RdPu")
# Third, using seaborn subplots for data visualisation
plt.figure(figsize = (20, 10))
sb.set(style = "whitegrid")
plt.subplot(2, 2, 1)
sb.distplot(data['SepalLengthCm'].values, bins = 30, kde = True, rug = True, color = "r").set(title = "Sepal Length")
plt.subplot(2, 2, 2)
sb.distplot(data['SepalWidthCm'].values, bins = 30, kde = True, rug = True, color = "b").set(title = "Sepal Width")
plt.subplot(2, 2, 3)
sb.distplot(data['PetalLengthCm'].values, bins = 30, kde = True, rug = True, color = "g").set(title = "Petal Length")
plt.subplot(2, 2, 4)
sb.distplot(data['PetalWidthCm'].values, bins = 30, kde = True, rug = True, color = "y").set(title = "Petal Width")
plt.show()
```
## Data Preprocessing ##
```
X = data.iloc[:, :-1].values
y = data.iloc[:, -1].values
print(X.shape)
print(y.shape)
print(data.shape)
# Splitting up of data into training and test set
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state = 0, test_size = 0.2, shuffle = True)
# shuffling of data is a good habit for creating a ML model
# Importing decision tree classifier
from sklearn.tree import DecisionTreeClassifier
dec = DecisionTreeClassifier()
dec.fit(X_train, y_train)
```
## Checking the accuracy of Decision Tree ##
```
# Importing the accuracy_score
from sklearn.metrics import accuracy_score
pred = dec.predict(X_test)
print(f"Accuracy : {accuracy_score(pred, y_test)* 100}")
```
Therefore, we got an immensely accurate Decision Tree scoring a 96.67 % of accuracy.
```
# Importing the classification report
from sklearn.metrics import classification_report
print(classification_report(y_test, pred))
```
In this report, we can clearly see the accuracy through f1-score which is 97 % (approx.). f1 - score = (2 * precision * recall / (precision + recall))
```
# Also, we can use seaborn confusion matrix
from sklearn.metrics import confusion_matrix
plt.figure(figsize=(8, 5))
sb.heatmap(confusion_matrix(y_test, pred), annot = True);
```
## Visualizing the Decision Tree ##
```
# Plotting Tree
from sklearn.tree import plot_tree
plt.figure(figsize = (10, 8))
tree = plot_tree(dec, feature_names = data.columns, class_names=data["Species"].unique().tolist(), precision = 4,
label = "all", filled = True)
plt.show()
```
# ASSIGNMENT COMPLETED #
|
github_jupyter
|
# Importing certain libraries
import pandas as pd
import numpy as np
import seaborn as sb
import matplotlib.pyplot as plt
%matplotlib inline
import sklearn.datasets as datasets
# Importing the dataset
data = pd.read_csv(r"D:\TSF\Task 4\Iris.csv")
# Displaying the dataset
data.head(151)
# For calculating the ideal number of clusters, we will unique functions to classify the number of species present
data["Species"].unique()
# Gathering the basic information
data.info()
# Data types
data.dtypes
# Checking for any null values or missing values
data.isnull().any()
# First, using seaborn pairplot for data visualisation
sb.set(style = "whitegrid")
plt.figure(figsize = (20, 10))
sb.pairplot(data, hue = "Species")
# Second, using seaborn heatmaps for data visualisation
sb.heatmap(data.corr(), annot = True, fmt = ".3g", linewidth = 0.5, linecolor = "Black", cmap = "RdPu")
# Third, using seaborn subplots for data visualisation
plt.figure(figsize = (20, 10))
sb.set(style = "whitegrid")
plt.subplot(2, 2, 1)
sb.distplot(data['SepalLengthCm'].values, bins = 30, kde = True, rug = True, color = "r").set(title = "Sepal Length")
plt.subplot(2, 2, 2)
sb.distplot(data['SepalWidthCm'].values, bins = 30, kde = True, rug = True, color = "b").set(title = "Sepal Width")
plt.subplot(2, 2, 3)
sb.distplot(data['PetalLengthCm'].values, bins = 30, kde = True, rug = True, color = "g").set(title = "Petal Length")
plt.subplot(2, 2, 4)
sb.distplot(data['PetalWidthCm'].values, bins = 30, kde = True, rug = True, color = "y").set(title = "Petal Width")
plt.show()
X = data.iloc[:, :-1].values
y = data.iloc[:, -1].values
print(X.shape)
print(y.shape)
print(data.shape)
# Splitting up of data into training and test set
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state = 0, test_size = 0.2, shuffle = True)
# shuffling of data is a good habit for creating a ML model
# Importing decision tree classifier
from sklearn.tree import DecisionTreeClassifier
dec = DecisionTreeClassifier()
dec.fit(X_train, y_train)
# Importing the accuracy_score
from sklearn.metrics import accuracy_score
pred = dec.predict(X_test)
print(f"Accuracy : {accuracy_score(pred, y_test)* 100}")
# Importing the classification report
from sklearn.metrics import classification_report
print(classification_report(y_test, pred))
# Also, we can use seaborn confusion matrix
from sklearn.metrics import confusion_matrix
plt.figure(figsize=(8, 5))
sb.heatmap(confusion_matrix(y_test, pred), annot = True);
# Plotting Tree
from sklearn.tree import plot_tree
plt.figure(figsize = (10, 8))
tree = plot_tree(dec, feature_names = data.columns, class_names=data["Species"].unique().tolist(), precision = 4,
label = "all", filled = True)
plt.show()
| 0.731442 | 0.962143 |
# Práctica 2: Procesamiento de Lenguaje Natural
## Parte 1: Análisis de sentimiento
##### Grupo Lab: 06 <br> Ismail Azizi González y Daniel Alfaro Miranda 3ºA
### Apartado a)
Primero leemos todas las reviews del archivo y configuramos dos vectores, uno con los datos y otro con 0s o 1s indicando si son buenas o malas reviews. Despues separamos los datos en entrenamiento y test, siendo 75% entrenamiento y 25% test.
```
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
import numpy as np
with open('yelp_labelled.txt') as f:
tmp = f.read().splitlines()
yelp_data = []
yelp_range = []
for i in range (0, len(tmp)):
tmp_data, tmp_range = tmp[i].strip().split('\t')
yelp_data.append(tmp_data)
yelp_range.append(tmp_range)
from sklearn.model_selection import train_test_split
train_data, test_data, train_target, test_target = train_test_split(yelp_data, yelp_range, test_size=0.25, shuffle = True, random_state=333)
```
### CountVectorizer(binary = True) sin TF/IDF solo monogramas
Configuramos el CountVectorizer para monogramas con binary = True, con la lista de palabras vacías de sklearn y sin TF/IDF
```
vectorizer = CountVectorizer(stop_words='english', binary = True)
train_vector_data=vectorizer.fit_transform(train_data)
test_vector_data=vectorizer.transform(test_data)
```
Utilizamos el clasificador Naive Bayes Multinomial, que es el más adecuado ya estamos sin TFIDF y por lo tanto solo tratamos la apariciones de las palabras como un número entero
```
from sklearn.naive_bayes import MultinomialNB
mnb_classifier = MultinomialNB()
mnb_classifier.fit(train_vector_data, train_target)
mnb_train_predictions = mnb_classifier.predict(train_vector_data)
mnb_test_predictions = mnb_classifier.predict(test_vector_data)
print("Multinomial Naive Bayes, porcentaje de aciertos en entrenamiento:"
, np.mean(mnb_train_predictions == train_target))
print("Multinomial Naive Bayes, porcentaje de aciertos en test:"
, np.mean(mnb_test_predictions == test_target))
from sklearn import tree
tree_classifier = tree.DecisionTreeClassifier(min_samples_leaf = 10)
tree_classifier.fit(train_vector_data, train_target)
tree_train_predictions = tree_classifier.predict(train_vector_data)
tree_test_predictions = tree_classifier.predict(test_vector_data)
print("Árbol, porcentaje de aciertos en entrenamiento:", np.mean(tree_train_predictions == train_target))
print("Árbol, porcentaje de aciertos en test:", np.mean(tree_test_predictions == test_target))
```
### CountVectorizer(binary = False) con TF/IDF solo monogramas
Configuramos el CountVectorizer para monogramas con binary = False, con la lista de palabras vacías de sklearn y usando TF/IDF
```
from sklearn.feature_extraction.text import TfidfTransformer
vectorizer2 = CountVectorizer(stop_words='english', binary = False)
train_vector_data_tfidf=vectorizer2.fit_transform(train_data)
test_vector_data_tfidf=vectorizer2.transform(test_data)
tfidfer = TfidfTransformer()
train_preprocessed = tfidfer.fit_transform(train_vector_data_tfidf)
test_preprocessed = tfidfer.transform(test_vector_data_tfidf)
```
Utilizamos el clasificador Naive Bayes Gaussiano, que es el más adecuado ya estamos usando TFIDF y por lo tanto hacemos la proporcion de las apariciones de los términos entre la frecuencia de las mismas en el documento, con lo cual podemos obtener valores discretos, los cuales se tratan mejor con el NBGaussiano
```
from sklearn.naive_bayes import GaussianNB
gnb_classifier = GaussianNB()
chunk_size=5
num_rows=len(train_target)
for i in range(0, (num_rows//chunk_size)):
train_chunk = train_preprocessed[i*chunk_size : (i+1)*chunk_size,:].toarray()
target_chunk = train_target[i*chunk_size : (i+1)*chunk_size]
gnb_classifier.partial_fit(train_chunk, target_chunk, classes=np.unique(train_target))
gnb_train_predictions=np.zeros_like(train_target)
gnb_test_predictions=np.zeros_like(test_target)
for i in range(0, (num_rows//chunk_size)):
train_chunk = train_preprocessed[i*chunk_size : (i+1)*chunk_size,:].toarray()
gnb_train_predictions[i*chunk_size : (i+1)*chunk_size] = gnb_classifier.predict(train_chunk)
num_rows=len(test_target)
for i in range(0, (num_rows//chunk_size)):
test_chunk = test_preprocessed[i*chunk_size : (i+1)*chunk_size,:].toarray()
gnb_test_predictions[i*chunk_size : (i+1)*chunk_size] = gnb_classifier.predict(test_chunk)
print("Gaussian Naive Bayes, porcentaje de aciertos en entrenamiento:", np.mean(gnb_train_predictions == train_target))
print("Gaussian Naive Bayes, porcentaje de aciertos en test:", np.mean(gnb_test_predictions == test_target))
tree_classifier2 = tree.DecisionTreeClassifier(min_samples_leaf = 10)
tree_classifier2.fit(train_preprocessed, train_target)
tree_train_predictions2 = tree_classifier2.predict(train_preprocessed)
tree_test_predictions2 = tree_classifier2.predict(test_preprocessed)
print("Árbol, porcentaje de aciertos en entrenamiento:", np.mean(tree_train_predictions2 == train_target))
print("Árbol, porcentaje de aciertos en test:", np.mean(tree_test_predictions2 == test_target))
```
### CountVectorizer(binary = True) sin TF/IDF, monogramas y bigramas
Configuramos el CountVectorizer para monogramas y bigramas con binary = True, con la lista de palabras vacías de sklearn y sin TF/IDF
```
vectorizer3 = CountVectorizer(stop_words='english', ngram_range=(1,2), binary = True)
train_vector_data_ngram_1_2=vectorizer3.fit_transform(train_data)
test_vector_data_ngram_1_2=vectorizer3.transform(test_data)
```
Usamos NBMultinomial igual que en el caso anterior sin TFIDF
```
mnb_classifier2 = MultinomialNB()
mnb_classifier2.fit(train_vector_data_ngram_1_2, train_target)
mnb_train_predictions2 = mnb_classifier2.predict(train_vector_data_ngram_1_2)
mnb_test_predictions2 = mnb_classifier2.predict(test_vector_data_ngram_1_2)
print("Multinomial Naive Bayes, porcentaje de aciertos en entrenamiento:"
, np.mean(mnb_train_predictions2 == train_target))
print("Multinomial Naive Bayes, porcentaje de aciertos en test:"
, np.mean(mnb_test_predictions2 == test_target))
tree_classifier3 = tree.DecisionTreeClassifier(min_samples_leaf = 10)
tree_classifier3.fit(train_vector_data_ngram_1_2, train_target)
tree_train_predictions3 = tree_classifier3.predict(train_vector_data_ngram_1_2)
tree_test_predictions3 = tree_classifier3.predict(test_vector_data_ngram_1_2)
print("Árbol, porcentaje de aciertos en entrenamiento:", np.mean(tree_train_predictions3 == train_target))
print("Árbol, porcentaje de aciertos en test:", np.mean(tree_test_predictions3 == test_target))
```
### CountVectorizer(binary = False) con TF/IDF, monogramas y bigramas
Configuramos el CountVectorizer para monogramas y bigramas con binary = False, con la lista de palabras vacías de sklearn y usando TF/IDF
```
vectorizer4 = CountVectorizer(stop_words='english', ngram_range=(1,2), binary = False)
train_vector_data_ngram_1_2_tfidf=vectorizer4.fit_transform(train_data)
test_vector_data_ngram_1_2_tfidf=vectorizer4.transform(test_data)
tfidfer2 = TfidfTransformer()
train_preprocessed2 = tfidfer2.fit_transform(train_vector_data_ngram_1_2_tfidf)
test_preprocessed2 = tfidfer2.fit_transform(test_vector_data_ngram_1_2_tfidf)
```
Usamos NBGaussiano igual que en el caso anterior con TFIDF
```
gnb_classifier2 = GaussianNB()
chunk_size2=5
num_rows2=len(train_target)
for i in range(0, (num_rows2//chunk_size2)):
train_chunk2 = train_preprocessed2[i*chunk_size2 : (i+1)*chunk_size2,:].toarray()
target_chunk2 = train_target[i*chunk_size2 : (i+1)*chunk_size2]
gnb_classifier2.partial_fit(train_chunk2, target_chunk2, classes=np.unique(train_target))
gnb_train_predictions2=np.zeros_like(train_target)
gnb_test_predictions2=np.zeros_like(test_target)
for i in range(0, (num_rows2//chunk_size2)):
train_chunk2 = train_preprocessed2[i*chunk_size2 : (i+1)*chunk_size2,:].toarray()
gnb_train_predictions2[i*chunk_size : (i+1)*chunk_size2] = gnb_classifier2.predict(train_chunk2)
num_rows2=len(test_target)
for i in range(0, (num_rows2//chunk_size2)):
test_chunk2 = test_preprocessed2[i*chunk_size2 : (i+1)*chunk_size2,:].toarray()
gnb_test_predictions2[i*chunk_size2 : (i+1)*chunk_size2] = gnb_classifier2.predict(test_chunk2)
print("Gaussian Naive Bayes, porcentaje de aciertos en entrenamiento:", np.mean(gnb_train_predictions2 == train_target))
print("Gaussian Naive Bayes, porcentaje de aciertos en test:", np.mean(gnb_test_predictions2 == test_target))
tree_classifier4 = tree.DecisionTreeClassifier(min_samples_leaf = 10)
tree_classifier4.fit(train_preprocessed2, train_target)
tree_train_predictions4 = tree_classifier4.predict(train_preprocessed2)
tree_test_predictions4 = tree_classifier4.predict(test_preprocessed2)
print("Árbol, porcentaje de aciertos en entrenamiento:", np.mean(tree_train_predictions4 == train_target))
print("Árbol, porcentaje de aciertos en test:", np.mean(tree_test_predictions4 == test_target))
```
## Análisis de los clasificadores
#### Clasificadores Naive Bayes Multinomiales para datos sin TF/IDF:
Tasa de aprendizaje muy alta.
Tasa de aciertos más alta de los clasificadores usados.
#### Clasificadores Naive Bayes Gaussianos para datos con TF/IDF
Tasa de aprendizaje muy alta.
Tasa de aciertos más baja de los clasificadores usados.
#### Árboles de decisión
Tasa de aprendizaje moderada.
Tasa de aciertos intermedia entre Gaussianos y Multinomiales.
Podemos deducir de lo anterior que los mejores clasificadores serían los árboles de decisión o los Naive Bayes Multinomiales, ya que los primeros tienen la tasa de aprendizaje considerablemente baja (70-75%) y una tasa de aciertos comparable a los otros clasificadores (alrededor del 72%) y los NBMultinomiales aunque tengan una tasa de aprendizaje más alta (>95%) también son los que más tasa de aciertos tienen de todos (alrededor del 75%).
Los árboles de decisión son buenos debido a que tal y como están diseñados, separan primero los nodos de tal forma que se discriminan las palabras que consigan mejor resultado en cada nodo (es decir, se reconocen muy bien las palabras que definen más claramente si la review es buena o mala).<br><br>
Los clasificadores Naive Bayes son sencillos y los que suelen usarse de referencia para otros clasificadores, aunque según en que contexto son buenos. En este caso los Multinomiales son mejores que los Gaussianos ya que añadir TF/IDF no aporta ninguna ventaja en esta situación, al reviews cortas y sin muchas palabras, no nos interesala proporcion de veces que aparecen las palabras, si no la presencia de estas sin más.<br><br>
Los bigramas a su vez no aportan nada concluyente, ya que nos interesa más fijarnos en palabras clave antes que en la probabilidad de una palabra condicionada por la aparicion de otra (por ejemplo, nos interesa más detectar palabras como "good", "bad", etc.. antes que intentar predecir que "good" va después de "very", ya que si tenemos en el conjunto de entrenamiento 7 apariciones de "good" después de "very", no ayuda nada a diferenciar que una review buena de una mala, ya que "bad" también puede ir detrás de "very").
### Variables con más poder discriminante
Arbol de decisión sobre el vectorizer sin TF/IDF y sin bigramas
```
def print_top25_features_in_trees(vectorizer, clf):
"""Prints features with the highest coefficient values"""
feature_names = vectorizer.get_feature_names()
top25 = np.argsort(clf.feature_importances_)[-25:]
reversed_top = top25[::-1]
print("Top 25 features in the tree\n")
print("%s" % ( " / ".join(feature_names[j] for j in reversed_top)))
print_top25_features_in_trees(vectorizer,tree_classifier)
```
Predominan las palabras de sentimiento positivo, ya que normalmente una review con una de esas palabras casi siempre va a ser buena, ya que en una review mala es muy raro que aparezcan palabras positivas y las reviews buenas suelen ser más genericas que las malas (en las malas muchas veces la queja es algo concreto, no simplemente se dice que es malo).<br><br>
Hay ruido, ya que hay palabras como "food" o "service" que por si solas pueden ser usadas en ambos tipos de review ("good/bad service" o "good/bad food")
### Variables con más presencia
NBMultinomial sobre el vectorizer sin TF/IDF y sin bigramas
```
def print_top25_features_per_class_in_NB(vectorizer, clf, class_labels):
"""Prints features with the highest coefficient values, per class"""
feature_names = vectorizer.get_feature_names()
print("Top 25 features per class\n")
for i, class_label in enumerate(class_labels):
if i >= 2:
break
top25 = np.argsort(clf.feature_log_prob_[i])[-25:]
reversed_top = top25[::-1]
print("%s: %s" % (class_label,
" / ".join(feature_names[j] for j in reversed_top)),'\n')
print_top25_features_per_class_in_NB(vectorizer,mnb_classifier,test_target)
```
Podemos ver que por lo general en las reviews buenas hay palabras como "good", "great", "delicious" que tienen bastante sentido para discriminar las reviews buenas y hay una cantidad bastante baja de ruido. Esto puede ser debido a, que como hemos remarcado antes, las reviews positivas suelen ser más genericas, ya que elogian algún aspecto mediante algún calificativo positivo.<br><br>
Las reviews malas por lo general tienen bastante más ruido, porque suelen quejarse de algo más concreto y no solo recurren al uso de calificativos negativos para ello (por ejemplo, aparece la palabra "eat" o "service" que no nos ayuda en nada a diferenciar ya que puede ser usada en ambos sentimientos). Si es verdad que algunas palabras como "bad" o "worst" aparecen, las cuales son bastante determinantes, pero por normal general hay mucho ruido.
### Apartado b)
#### Naive Bayes
```
from sklearn.metrics import classification_report, confusion_matrix
classifier=mnb_classifier
predictions = mnb_test_predictions
print(classification_report(test_target, predictions))
```
Tiene ligeramente mejor precision para las reviews negativas que para las positivas, pero recupera más reviews positivas que negativas.
#### Arbol
```
classifier=tree_classifier
predictions = tree_test_predictions
print(classification_report(test_target, predictions))
```
Recupera la mayoría de opiniones negativas a costa de tener una precision considerablemente baja, y tiene una muy buena precision a la hora de recuperar opiniones positivas a costa de solo recuperar poco más de la mitad.
### Conclusiones
El clasificador de arbol acierta un 10% más en las opiniones positivas, pero a costa de recuperar un 24% menos de estas en comparacion con Naive Bayes Multinomial.<br>
En cuanto a las opiniones negativas, el clasificador de árbol recupera un 88% de ellas (18% más que el Naive Bayes) pero a costa de acertar solo un 67%, mientras que Naive Bayes acierta un 11% más de las veces.<br><br>
De estos datos podemos deducir que Naive Bayes es más consistente en su labor de clasificar que el clasificador de arbol.
### Arbol de decision representado
```
from sklearn.tree import plot_tree
import matplotlib.pyplot as plt
plt.figure(figsize=(15,15))
plot_tree(tree_classifier, filled=True, rounded=True, max_depth = 8,
feature_names = vectorizer.get_feature_names(), class_names = test_target)
plt.show()
```
El árbol está totalmente inclinado hacia la izquierda, ya que a la derecha separa muy rápido las opiniones positivas de las negativas. Se puede observar la precision del 83% en las opiniones positivas en las ramas derechas del árbol, ya que clasifica muy rapido las opiniones positivas (a costa de llevarse consigo algunas opiniones negativas y clasificarlas como positivas). Por lo general, en las primeras 8 alturas del árbol no se nota tanto la presencia de nodos impuros en la rama derecha, sin embargo la rama izquierda sí tiene muchos nodos impuros, ya que tarda en diferenciar las opiniones negativas.
### Ejemplo de falsos positivos y falsos negativos
#### Falsos positivos NB
```
j = 0
for i in range(0, 250):
if test_target[i] == '0' and mnb_test_predictions[i] == '1':
print(test_data[i])
print("\n")
j = j + 1
if j == 2:
break
```
La primera review ha sido calificada como positiva dado que tiene la palabra "Vegas" (sabemos que es esta palabra porque es una de las más discriminantes para determinar una review buena, aunque solo es ruido), la cual está presente en 22 reviews y de esas 22, 15 son positivas. El clasificador ha utilizado una palabra que no tiene nada que ver con que sea una review buena o mala para clasificar la review. En este caso, la palabra solo es ruido pero aun así ha conseguido confundir al clasificador.<br><br>
Con la segunda review ocurre lo mismo, la palabra "place" es una de las más discriminantes para determinar una review buena, pero solamente es ruido y no debería definir nada, ya que puede ser usada en ambos contextos. Hay 112 apariciones de "place" y la mayoría son en reviews positivas, por eso el clasificador piensa que sirve para determinar el sentimiento de la review
#### Falsos negativos NB
```
j = 0
for i in range(0, 250):
if test_target[i] == '1' and mnb_test_predictions[i] == '0':
if j == 11 or j == 13:
print(test_data[i])
print("\n")
j = j + 1
if j == 15:
break
```
En estos dos casos hemos cogido dos frases con las palabras "wait" y "dissapointed" que en principio debería denotar que la review es mala (no es bueno esperar y disappointed significa decepcionado) pero dichas palabras quieren referirse a aspectos positivos, ya que antes de dichas palabras hay una negación. Por ese motivo el clasificador se ha equivocado, porque no ha conseguido entender el contexto de la frase y solo se ha fijado en la palabra en concreto.
#### Falsos positivos arbol
```
j = 0
for i in range(0, 250):
if test_target[i] == '0' and tree_test_predictions[i] == '1':
print(test_data[i])
print("\n")
j = j + 1
if j == 2:
break
```
En este caso hemos tenido el mismo problema que antes con la palabra "Vegas", y en la segunda review hemos tenido un problema con la palabra "good", que se utiliza para reviews buenas normalmente, pero otra vez el clasificador no ha entendido el contexto de la frase que niega la palabra "good"
#### Falsos negativos arbol
```
j = 0
for i in range(0, 250):
if test_target[i] == '1' and tree_test_predictions[i] == '0':
if j == 25 or j == 38:
print(test_data[i])
print("\n")
j = j + 1
if j == 39:
break
```
En este caso hemos cogido dos reviews donde aparecen las palabras "food" y "place" presentes tanto en reviews buenas como malas, con lo cual no debería valer para diferenciar reviews buenas de malas, pero al tener tantas apariciones, se utiliza en el arbol de decision para partir un nodo, y si luego el nodo no se vuelve a dividir o se divide en base a otra palabra que no figure en una de las frases que contenia las palabras anteriores, puede quedarse la frase perdida en un nodo impuro.
### Mejorar el clasificador
Para mejorar el clasificador podriamos hacer que se fijase en el contexto de la frase y no solo en palabras sueltas. Si lo tomamos desde el punto de vista más sintáctico, podríamos analizar no solo los adjetivos que describen algo bueno o algo malo, si no también los cuantificadores o modificadores que pueden cambiar el sentido del adjetivo.
|
github_jupyter
|
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
import numpy as np
with open('yelp_labelled.txt') as f:
tmp = f.read().splitlines()
yelp_data = []
yelp_range = []
for i in range (0, len(tmp)):
tmp_data, tmp_range = tmp[i].strip().split('\t')
yelp_data.append(tmp_data)
yelp_range.append(tmp_range)
from sklearn.model_selection import train_test_split
train_data, test_data, train_target, test_target = train_test_split(yelp_data, yelp_range, test_size=0.25, shuffle = True, random_state=333)
vectorizer = CountVectorizer(stop_words='english', binary = True)
train_vector_data=vectorizer.fit_transform(train_data)
test_vector_data=vectorizer.transform(test_data)
from sklearn.naive_bayes import MultinomialNB
mnb_classifier = MultinomialNB()
mnb_classifier.fit(train_vector_data, train_target)
mnb_train_predictions = mnb_classifier.predict(train_vector_data)
mnb_test_predictions = mnb_classifier.predict(test_vector_data)
print("Multinomial Naive Bayes, porcentaje de aciertos en entrenamiento:"
, np.mean(mnb_train_predictions == train_target))
print("Multinomial Naive Bayes, porcentaje de aciertos en test:"
, np.mean(mnb_test_predictions == test_target))
from sklearn import tree
tree_classifier = tree.DecisionTreeClassifier(min_samples_leaf = 10)
tree_classifier.fit(train_vector_data, train_target)
tree_train_predictions = tree_classifier.predict(train_vector_data)
tree_test_predictions = tree_classifier.predict(test_vector_data)
print("Árbol, porcentaje de aciertos en entrenamiento:", np.mean(tree_train_predictions == train_target))
print("Árbol, porcentaje de aciertos en test:", np.mean(tree_test_predictions == test_target))
from sklearn.feature_extraction.text import TfidfTransformer
vectorizer2 = CountVectorizer(stop_words='english', binary = False)
train_vector_data_tfidf=vectorizer2.fit_transform(train_data)
test_vector_data_tfidf=vectorizer2.transform(test_data)
tfidfer = TfidfTransformer()
train_preprocessed = tfidfer.fit_transform(train_vector_data_tfidf)
test_preprocessed = tfidfer.transform(test_vector_data_tfidf)
from sklearn.naive_bayes import GaussianNB
gnb_classifier = GaussianNB()
chunk_size=5
num_rows=len(train_target)
for i in range(0, (num_rows//chunk_size)):
train_chunk = train_preprocessed[i*chunk_size : (i+1)*chunk_size,:].toarray()
target_chunk = train_target[i*chunk_size : (i+1)*chunk_size]
gnb_classifier.partial_fit(train_chunk, target_chunk, classes=np.unique(train_target))
gnb_train_predictions=np.zeros_like(train_target)
gnb_test_predictions=np.zeros_like(test_target)
for i in range(0, (num_rows//chunk_size)):
train_chunk = train_preprocessed[i*chunk_size : (i+1)*chunk_size,:].toarray()
gnb_train_predictions[i*chunk_size : (i+1)*chunk_size] = gnb_classifier.predict(train_chunk)
num_rows=len(test_target)
for i in range(0, (num_rows//chunk_size)):
test_chunk = test_preprocessed[i*chunk_size : (i+1)*chunk_size,:].toarray()
gnb_test_predictions[i*chunk_size : (i+1)*chunk_size] = gnb_classifier.predict(test_chunk)
print("Gaussian Naive Bayes, porcentaje de aciertos en entrenamiento:", np.mean(gnb_train_predictions == train_target))
print("Gaussian Naive Bayes, porcentaje de aciertos en test:", np.mean(gnb_test_predictions == test_target))
tree_classifier2 = tree.DecisionTreeClassifier(min_samples_leaf = 10)
tree_classifier2.fit(train_preprocessed, train_target)
tree_train_predictions2 = tree_classifier2.predict(train_preprocessed)
tree_test_predictions2 = tree_classifier2.predict(test_preprocessed)
print("Árbol, porcentaje de aciertos en entrenamiento:", np.mean(tree_train_predictions2 == train_target))
print("Árbol, porcentaje de aciertos en test:", np.mean(tree_test_predictions2 == test_target))
vectorizer3 = CountVectorizer(stop_words='english', ngram_range=(1,2), binary = True)
train_vector_data_ngram_1_2=vectorizer3.fit_transform(train_data)
test_vector_data_ngram_1_2=vectorizer3.transform(test_data)
mnb_classifier2 = MultinomialNB()
mnb_classifier2.fit(train_vector_data_ngram_1_2, train_target)
mnb_train_predictions2 = mnb_classifier2.predict(train_vector_data_ngram_1_2)
mnb_test_predictions2 = mnb_classifier2.predict(test_vector_data_ngram_1_2)
print("Multinomial Naive Bayes, porcentaje de aciertos en entrenamiento:"
, np.mean(mnb_train_predictions2 == train_target))
print("Multinomial Naive Bayes, porcentaje de aciertos en test:"
, np.mean(mnb_test_predictions2 == test_target))
tree_classifier3 = tree.DecisionTreeClassifier(min_samples_leaf = 10)
tree_classifier3.fit(train_vector_data_ngram_1_2, train_target)
tree_train_predictions3 = tree_classifier3.predict(train_vector_data_ngram_1_2)
tree_test_predictions3 = tree_classifier3.predict(test_vector_data_ngram_1_2)
print("Árbol, porcentaje de aciertos en entrenamiento:", np.mean(tree_train_predictions3 == train_target))
print("Árbol, porcentaje de aciertos en test:", np.mean(tree_test_predictions3 == test_target))
vectorizer4 = CountVectorizer(stop_words='english', ngram_range=(1,2), binary = False)
train_vector_data_ngram_1_2_tfidf=vectorizer4.fit_transform(train_data)
test_vector_data_ngram_1_2_tfidf=vectorizer4.transform(test_data)
tfidfer2 = TfidfTransformer()
train_preprocessed2 = tfidfer2.fit_transform(train_vector_data_ngram_1_2_tfidf)
test_preprocessed2 = tfidfer2.fit_transform(test_vector_data_ngram_1_2_tfidf)
gnb_classifier2 = GaussianNB()
chunk_size2=5
num_rows2=len(train_target)
for i in range(0, (num_rows2//chunk_size2)):
train_chunk2 = train_preprocessed2[i*chunk_size2 : (i+1)*chunk_size2,:].toarray()
target_chunk2 = train_target[i*chunk_size2 : (i+1)*chunk_size2]
gnb_classifier2.partial_fit(train_chunk2, target_chunk2, classes=np.unique(train_target))
gnb_train_predictions2=np.zeros_like(train_target)
gnb_test_predictions2=np.zeros_like(test_target)
for i in range(0, (num_rows2//chunk_size2)):
train_chunk2 = train_preprocessed2[i*chunk_size2 : (i+1)*chunk_size2,:].toarray()
gnb_train_predictions2[i*chunk_size : (i+1)*chunk_size2] = gnb_classifier2.predict(train_chunk2)
num_rows2=len(test_target)
for i in range(0, (num_rows2//chunk_size2)):
test_chunk2 = test_preprocessed2[i*chunk_size2 : (i+1)*chunk_size2,:].toarray()
gnb_test_predictions2[i*chunk_size2 : (i+1)*chunk_size2] = gnb_classifier2.predict(test_chunk2)
print("Gaussian Naive Bayes, porcentaje de aciertos en entrenamiento:", np.mean(gnb_train_predictions2 == train_target))
print("Gaussian Naive Bayes, porcentaje de aciertos en test:", np.mean(gnb_test_predictions2 == test_target))
tree_classifier4 = tree.DecisionTreeClassifier(min_samples_leaf = 10)
tree_classifier4.fit(train_preprocessed2, train_target)
tree_train_predictions4 = tree_classifier4.predict(train_preprocessed2)
tree_test_predictions4 = tree_classifier4.predict(test_preprocessed2)
print("Árbol, porcentaje de aciertos en entrenamiento:", np.mean(tree_train_predictions4 == train_target))
print("Árbol, porcentaje de aciertos en test:", np.mean(tree_test_predictions4 == test_target))
def print_top25_features_in_trees(vectorizer, clf):
"""Prints features with the highest coefficient values"""
feature_names = vectorizer.get_feature_names()
top25 = np.argsort(clf.feature_importances_)[-25:]
reversed_top = top25[::-1]
print("Top 25 features in the tree\n")
print("%s" % ( " / ".join(feature_names[j] for j in reversed_top)))
print_top25_features_in_trees(vectorizer,tree_classifier)
def print_top25_features_per_class_in_NB(vectorizer, clf, class_labels):
"""Prints features with the highest coefficient values, per class"""
feature_names = vectorizer.get_feature_names()
print("Top 25 features per class\n")
for i, class_label in enumerate(class_labels):
if i >= 2:
break
top25 = np.argsort(clf.feature_log_prob_[i])[-25:]
reversed_top = top25[::-1]
print("%s: %s" % (class_label,
" / ".join(feature_names[j] for j in reversed_top)),'\n')
print_top25_features_per_class_in_NB(vectorizer,mnb_classifier,test_target)
from sklearn.metrics import classification_report, confusion_matrix
classifier=mnb_classifier
predictions = mnb_test_predictions
print(classification_report(test_target, predictions))
classifier=tree_classifier
predictions = tree_test_predictions
print(classification_report(test_target, predictions))
from sklearn.tree import plot_tree
import matplotlib.pyplot as plt
plt.figure(figsize=(15,15))
plot_tree(tree_classifier, filled=True, rounded=True, max_depth = 8,
feature_names = vectorizer.get_feature_names(), class_names = test_target)
plt.show()
j = 0
for i in range(0, 250):
if test_target[i] == '0' and mnb_test_predictions[i] == '1':
print(test_data[i])
print("\n")
j = j + 1
if j == 2:
break
j = 0
for i in range(0, 250):
if test_target[i] == '1' and mnb_test_predictions[i] == '0':
if j == 11 or j == 13:
print(test_data[i])
print("\n")
j = j + 1
if j == 15:
break
j = 0
for i in range(0, 250):
if test_target[i] == '0' and tree_test_predictions[i] == '1':
print(test_data[i])
print("\n")
j = j + 1
if j == 2:
break
j = 0
for i in range(0, 250):
if test_target[i] == '1' and tree_test_predictions[i] == '0':
if j == 25 or j == 38:
print(test_data[i])
print("\n")
j = j + 1
if j == 39:
break
| 0.312895 | 0.942295 |
# Install
### References
* Official
* wiki
* https://github.com/BVLC/caffe/wiki
* Homepage
* http://caffe.berkeleyvision.org/
* Others
* https://hpc.uiowa.edu/sites/hpc.uiowa.edu/files/wysiwyg_uploads/ModifiedCaffeTutorial_0.pdf
### Prerequirements
* DB (http://caffe.berkeleyvision.org/tutorial/data.html)
* level db
* fast key-value storage library written at Google
* https://github.com/google/leveldb
* lmdb
* lightning memory-mapped database
* https://en.wikipedia.org/wiki/Lightning_Memory-Mapped_Database
* protobuf
* for serialize/deserialize
* https://en.wikipedia.org/wiki/Protocol_Buffers
* hdf5
* file format, Hierarchical Data Format
* https://en.wikipedia.org/wiki/Hierarchical_Data_Format
* glog
* logging library from google
* https://github.com/google/glog
* gflags
* command line flag parser
* https://github.com/gflags/gflags
* for python
* for pygraphviz, which pydot is dependent on
* PyGraphviz doesn't work without Graphviz
* `sudo apt-get install graphviz`
* `pip3 install pydot scikit-image pytest`
```sh
sudo apt-get install libprotobuf-dev libleveldb-dev libsnappy-dev libhdf5-serial-dev protobuf-compiler
sudo apt-get install --no-install-recommends libboost-all-dev
sudo apt-get install libatlas-base-dev
sudo apt-get install libgflags-dev libgoogle-glog-dev liblmdb-dev
```
### Fix
```diff
diff --git a/Makefile b/Makefile
index 4d32416..ad173f8 100644
--- a/Makefile
+++ b/Makefile
@@ -178,7 +178,7 @@ ifneq ($(CPU_ONLY), 1)
LIBRARIES := cudart cublas curand
endif
-LIBRARIES += glog gflags protobuf boost_system boost_filesystem m hdf5_hl hdf5
+LIBRARIES += glog gflags protobuf boost_system boost_filesystem m hdf5_serial_hl hdf5_serial
# handle IO dependencies
USE_LEVELDB ?= 1
```
### Configurations
#### Makefile.config
* for Multi-GPU
```
USE_NCCL := 1
```
* install nccl (http://www.nvidia.com/object/caffe-installation.html)
```
$ git clone https://github.com/NVIDIA/nccl.git
$ cd nccl
$ sudo make install -j4
```
* for cudnn
```
USE_CUDNN:=1
```
* for opencv
```
OPENCV_VERSION:=3
```
* for python3
```
PYTHON_LIBRARIES := boost_python-py35 python3.5m
PYTHON_INCLUDE := /usr/include/python3.5m \
/usr/lib/python3.5/dist-packages/numpy/core/include
```
* Custumize include and library dir
* e.g. I installed OpenCV with `prefix=/opt/` so I need `/opt/include`, `/opt/lib`
```
INCLUDE_DIRS := $(PYTHON_INCLUDE) /usr/local/include /usr/include/hdf5/serial/ /opt/include/
LIBRARY_DIRS := $(PYTHON_LIB) /usr/local/lib /usr/lib /opt/lib
```
#### Makefile
* for python3
```diff
@@ -533,7 +533,7 @@ runtest: $(TEST_ALL_BIN)
$(TEST_ALL_BIN) $(TEST_GPUID) --gtest_shuffle $(TEST_FILTER)
pytest: py
- cd python; python -m unittest discover -s caffe/test
+ cd python; python3 -m unittest discover -s caffe/test
mattest: mat
cd matlab; $(MATLAB_DIR)/bin/matlab -nodisplay -r 'caffe.run_tests(), exit()'
```
### How to run my own code
* ```export PYTHONPATH=~/work/caffe/ssd/python/```
* Easy way
* put a **single** `*.cpp` file in ./examples
* Build own project by linking caffe.so???
* 아직 모른다.
### save/load caffemodel
* save
* declared in solver.prototxt
```
# snapshot intermediate results
snapshot: 5000
snapshot_prefix: "examples/mnist/lenet"
```
* you can see `*.caffemodel`, `*.solverstate`
```
-rw-rw-r-- 1 rofox rofox 1725006 May 25 10:59 lenet_iter_10000.caffemodel
-rw-rw-r-- 1 rofox rofox 1724471 May 25 10:59 lenet_iter_10000.solverstate
-rw-rw-r-- 1 rofox rofox 1725006 May 25 10:58 lenet_iter_5000.caffemodel
-rw-rw-r-- 1 rofox rofox 1724470 May 25 10:58 lenet_iter_5000.solverstate
```
* load
*
### Multi GPU Test
* cifar10 test total batch 100
| 1 gpu | 2 gpu | 4 gpu
:-:|:-:|:-:|:-:
local_batch x num_gpus| 100x1 | 50x2 | 25x4
performance (iters/sec)| 30.2061 | 49.4755 | 57.3761
ratio| 1 | 1.6379 | 1.8994
|
github_jupyter
|
sudo apt-get install libprotobuf-dev libleveldb-dev libsnappy-dev libhdf5-serial-dev protobuf-compiler
sudo apt-get install --no-install-recommends libboost-all-dev
sudo apt-get install libatlas-base-dev
sudo apt-get install libgflags-dev libgoogle-glog-dev liblmdb-dev
diff --git a/Makefile b/Makefile
index 4d32416..ad173f8 100644
--- a/Makefile
+++ b/Makefile
@@ -178,7 +178,7 @@ ifneq ($(CPU_ONLY), 1)
LIBRARIES := cudart cublas curand
endif
-LIBRARIES += glog gflags protobuf boost_system boost_filesystem m hdf5_hl hdf5
+LIBRARIES += glog gflags protobuf boost_system boost_filesystem m hdf5_serial_hl hdf5_serial
# handle IO dependencies
USE_LEVELDB ?= 1
USE_NCCL := 1
$ git clone https://github.com/NVIDIA/nccl.git
$ cd nccl
$ sudo make install -j4
USE_CUDNN:=1
OPENCV_VERSION:=3
PYTHON_LIBRARIES := boost_python-py35 python3.5m
PYTHON_INCLUDE := /usr/include/python3.5m \
/usr/lib/python3.5/dist-packages/numpy/core/include
INCLUDE_DIRS := $(PYTHON_INCLUDE) /usr/local/include /usr/include/hdf5/serial/ /opt/include/
LIBRARY_DIRS := $(PYTHON_LIB) /usr/local/lib /usr/lib /opt/lib
@@ -533,7 +533,7 @@ runtest: $(TEST_ALL_BIN)
$(TEST_ALL_BIN) $(TEST_GPUID) --gtest_shuffle $(TEST_FILTER)
pytest: py
- cd python; python -m unittest discover -s caffe/test
+ cd python; python3 -m unittest discover -s caffe/test
mattest: mat
cd matlab; $(MATLAB_DIR)/bin/matlab -nodisplay -r 'caffe.run_tests(), exit()'
* Easy way
* put a **single** `*.cpp` file in ./examples
* Build own project by linking caffe.so???
* 아직 모른다.
### save/load caffemodel
* save
* declared in solver.prototxt
* you can see `*.caffemodel`, `*.solverstate`
| 0.270288 | 0.574544 |
# Оптимизация многомерной функции с ограничениями
```
import numpy as np
import scipy.optimize as sopt
import matplotlib.pyplot as plt
%matplotlib inline
# точки в вершинах треугольной области допустимых значений параметров
H = 100.
pts = np.array([[2., 1.],
[H-2.,1.],
[H/2.,H/2.-1.]])
def closeBorder(nodes):
return np.vstack((nodes, nodes[0]))
brd = closeBorder(pts)
# целевая функция
def G(L, R, H, lst=[]):
lst.append([L,R])
return H*(L-R**2)**2+(1.-L)**2
# расчет целевой функции на прямоугольной равномерной решетке
x = np.linspace(pts[:,0].min(), pts[:,0].max(), 100)
y = np.linspace(pts[:,1].min(), pts[:,1].max(), 100)
X, Y = np.meshgrid(x, y)
Z = G(X, Y, H)
# контурная карта целевой функции
#plt.figure(figsize=(10,10))
plt.contourf(X, Y, np.log(Z+1))
plt.plot(brd[:,0], brd[:,1])
#plt.xticks(range(5))
#plt.xlim(0, 5)
#plt.ylim(0, 5)
plt.colorbar()
# [1., 1-H, 1]
# [np.inf, np.inf, np.inf]
# [[1., -1], [-1, -1], [0, 1]]
lc = sopt.LinearConstraint([[1., -1], [-1, -1], [0, 1]],
[1., 1-H, 1],
[np.inf, np.inf, np.inf])
# 1 <= 1*L -1*R <= np.inf
# 1 - H <= -1*L -1*R <= np.inf
# 1 <= 0*L +1*R <= np.inf
```
### Оптимизация функции G с ограничениями в виде неравенств методом COBYLA
```
ineq_constr = {'type':'ineq',
# ограничения в виде неравенств (правая часть неравенств c(x)>=0)
'fun':lambda x : np.array([-1.+x[0]-x[1],
H-1.-x[0]-x[1],
-1.+x[1]]),
# якобиан 'fun'
'jac':lambda x : np.array([[ 1., -1.],
[-1., -1.],
[ 0., 1.]])
}
lst = []
ret = sopt.minimize(lambda x, H, lst: G(x[0], x[1], H, lst), [80., 10], args=(H, lst),
method='COBYLA',
options={'tol': 1e-12, 'disp': True, 'maxiter':100000},
constraints=[ineq_constr])
ret
arr = np.array(lst)
arr.shape
plt.figure(figsize=(10,10))
plt.contourf(X, Y, np.log(Z+1))
plt.plot(brd[:,0], brd[:,1])
plt.plot(arr[:,0], arr[:,1])
plt.plot(ret.x[0], ret.x[1], 'o')
#plt.xticks(range(5))
#plt.xlim(0, 5)
#plt.ylim(0, 5)
plt.colorbar()
```
### Оптимизация модифицированной функции G без ограничений методом Нелдера-Мида (деформируемого многогранника)
```
def G(L, R, H, lst=[]):
lst.append([L,R])
if (-1.+L-R >= 0) and (H-1.-L-R>=0) and (-1.+R>=0):
return H*(L-R**2)**2+(1.-L)**2
else:
return 1e15
lst = []
ret = sopt.minimize(lambda x, H, lst: G(x[0], x[1], H, lst), [80., 10], args=(H, lst),
method='Nelder-Mead',
options={'xatol': 1e-9, 'disp': True, 'maxiter':100000})
arr = np.array(lst)
plt.figure(figsize=(10,10))
plt.contourf(X, Y, np.log(Z+1))
plt.plot(brd[:,0], brd[:,1])
plt.plot(arr[:,0], arr[:,1])
plt.plot(ret.x[0], ret.x[1], 'o')
#plt.xticks(range(5))
#plt.xlim(0, 5)
#plt.ylim(0, 5)
#plt.axis('equal')
plt.colorbar()
ret
```
|
github_jupyter
|
import numpy as np
import scipy.optimize as sopt
import matplotlib.pyplot as plt
%matplotlib inline
# точки в вершинах треугольной области допустимых значений параметров
H = 100.
pts = np.array([[2., 1.],
[H-2.,1.],
[H/2.,H/2.-1.]])
def closeBorder(nodes):
return np.vstack((nodes, nodes[0]))
brd = closeBorder(pts)
# целевая функция
def G(L, R, H, lst=[]):
lst.append([L,R])
return H*(L-R**2)**2+(1.-L)**2
# расчет целевой функции на прямоугольной равномерной решетке
x = np.linspace(pts[:,0].min(), pts[:,0].max(), 100)
y = np.linspace(pts[:,1].min(), pts[:,1].max(), 100)
X, Y = np.meshgrid(x, y)
Z = G(X, Y, H)
# контурная карта целевой функции
#plt.figure(figsize=(10,10))
plt.contourf(X, Y, np.log(Z+1))
plt.plot(brd[:,0], brd[:,1])
#plt.xticks(range(5))
#plt.xlim(0, 5)
#plt.ylim(0, 5)
plt.colorbar()
# [1., 1-H, 1]
# [np.inf, np.inf, np.inf]
# [[1., -1], [-1, -1], [0, 1]]
lc = sopt.LinearConstraint([[1., -1], [-1, -1], [0, 1]],
[1., 1-H, 1],
[np.inf, np.inf, np.inf])
# 1 <= 1*L -1*R <= np.inf
# 1 - H <= -1*L -1*R <= np.inf
# 1 <= 0*L +1*R <= np.inf
ineq_constr = {'type':'ineq',
# ограничения в виде неравенств (правая часть неравенств c(x)>=0)
'fun':lambda x : np.array([-1.+x[0]-x[1],
H-1.-x[0]-x[1],
-1.+x[1]]),
# якобиан 'fun'
'jac':lambda x : np.array([[ 1., -1.],
[-1., -1.],
[ 0., 1.]])
}
lst = []
ret = sopt.minimize(lambda x, H, lst: G(x[0], x[1], H, lst), [80., 10], args=(H, lst),
method='COBYLA',
options={'tol': 1e-12, 'disp': True, 'maxiter':100000},
constraints=[ineq_constr])
ret
arr = np.array(lst)
arr.shape
plt.figure(figsize=(10,10))
plt.contourf(X, Y, np.log(Z+1))
plt.plot(brd[:,0], brd[:,1])
plt.plot(arr[:,0], arr[:,1])
plt.plot(ret.x[0], ret.x[1], 'o')
#plt.xticks(range(5))
#plt.xlim(0, 5)
#plt.ylim(0, 5)
plt.colorbar()
def G(L, R, H, lst=[]):
lst.append([L,R])
if (-1.+L-R >= 0) and (H-1.-L-R>=0) and (-1.+R>=0):
return H*(L-R**2)**2+(1.-L)**2
else:
return 1e15
lst = []
ret = sopt.minimize(lambda x, H, lst: G(x[0], x[1], H, lst), [80., 10], args=(H, lst),
method='Nelder-Mead',
options={'xatol': 1e-9, 'disp': True, 'maxiter':100000})
arr = np.array(lst)
plt.figure(figsize=(10,10))
plt.contourf(X, Y, np.log(Z+1))
plt.plot(brd[:,0], brd[:,1])
plt.plot(arr[:,0], arr[:,1])
plt.plot(ret.x[0], ret.x[1], 'o')
#plt.xticks(range(5))
#plt.xlim(0, 5)
#plt.ylim(0, 5)
#plt.axis('equal')
plt.colorbar()
ret
| 0.223123 | 0.925567 |
<a href="https://colab.research.google.com/github/DarkTitan007/Data_Cleaning_NLP/blob/main/Untitled2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import pandas as pd
import numpy as np
from nltk.tokenize import word_tokenize
from nltk import pos_tag
from nltk.corpus import stopwords
from nltk.stem import WordNetLemmatizer
from sklearn.preprocessing import LabelEncoder
from collections import defaultdict
from nltk.corpus import wordnet as wn
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn import model_selection, naive_bayes, svm
from sklearn.metrics import accuracy_score
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
import matplotlib.pyplot as plt
import seaborn as sns
np.random.seed(500)
Corpus = pd.read_csv(r"/amit.csv",encoding='latin-1')
Corpus.head()
Corpus.info()
sns.countplot(Corpus.category)
plt.xlabel('Category')
plt.title('CountPlot')
import nltk
nltk.download('punkt')
nltk.download('wordnet')
nltk.download('averaged_perceptron_tagger')
nltk.download('stopwords')
# 1. Removing Blank Spaces
Corpus['text'].dropna(inplace=True)
# 2. Changing all text to lowercase
Corpus['text_original'] = Corpus['text']
Corpus['text'] = str(Corpus['text'])
Corpus['text'] = [entry.lower() for entry in Corpus['text']]
# 3. Tokenization-In this each entry in the corpus will be broken into set of words
Corpus['text']= [word_tokenize(entry) for entry in Corpus['text']]
# 4. Remove Stop words, Non-Numeric and perfoming Word Stemming/Lemmenting.
# WordNetLemmatizer requires Pos tags to understand if the word is noun or verb or adjective etc. By default it is set to Noun
tag_map = defaultdict(lambda : wn.NOUN)
tag_map['J'] = wn.ADJ
tag_map['V'] = wn.VERB
tag_map['R'] = wn.ADV
Corpus.head()
for index,entry in enumerate(Corpus['text']):
# Declaring Empty List to store the words that follow the rules for this step
Final_words = []
# Initializing WordNetLemmatizer()
word_Lemmatized = WordNetLemmatizer()
# pos_tag function below will provide the 'tag' i.e if the word is Noun(N) or Verb(V) or something else.
for word, tag in pos_tag(entry):
# Below condition is to check for Stop words and consider only alphabets
if word not in stopwords.words('english') and word.isalpha():
word_Final = word_Lemmatized.lemmatize(word,tag_map[tag[0]])
Final_words.append(word_Final)
# The final processed set of words for each iteration will be stored in 'text_final'
Corpus.loc[index,'text_final'] = str(Final_words)
Corpus.drop(['text'], axis=1)
output_path = 'preprocessed_data.csv'
Corpus.to_csv(output_path, index=False)
Train_X, Test_X, Train_Y, Test_Y = model_selection.train_test_split(Corpus['text_final'],Corpus['category'],test_size=0.3)
Encoder = LabelEncoder()
Train_Y = Encoder.fit_transform(Train_Y)
Test_Y = Encoder.fit_transform(Test_Y)
Tfidf_vect = TfidfVectorizer(max_features=5000)
Tfidf_vect.fit(Corpus['text_final'])
Train_X_Tfidf = Tfidf_vect.transform(Train_X)
Test_X_Tfidf = Tfidf_vect.transform(Test_X)
print(Tfidf_vect.vocabulary_)
print(Train_X_Tfidf)
# fit the training dataset on the NB classifier
Naive = naive_bayes.MultinomialNB()
Naive.fit(Train_X_Tfidf,Train_Y)
# predict the labels on validation dataset
predictions_NB = Naive.predict(Test_X_Tfidf)
# Use accuracy_score function to get the accuracy
print("Naive Bayes Accuracy Score -> ",accuracy_score(predictions_NB, Test_Y)*100)
from sklearn import model_selection, naive_bayes, svm
from sklearn.metrics import accuracy_score
Naive = naive_bayes.MultinomialNB()
Naive.fit(Train_X_Tfidf,Train_Y)
# predict the labels on validation dataset
predictions_NB = Naive.predict(Test_X_Tfidf)
# Use accuracy_score function to get the accuracy
print("Naive Bayes Accuracy Score -> ",accuracy_score(predictions_NB, Test_Y)*100)
# Classifier - Algorithm - SVM
# fit the training dataset on the classifier
SVM = svm.SVC(C=1.0, kernel='linear', degree=3, gamma='auto')
SVM.fit(Train_X_Tfidf,Train_Y)
# predict the labels on validation dataset
predictions_SVM = SVM.predict(Test_X_Tfidf)
Train_X_Tfidf.shape
print(classification_report(Test_Y, predictions_NB))
# Classifier - Algorithm - SVM
# fit the training dataset on the classifier
SVM = svm.SVC(C=1.0, kernel='linear', degree=3, gamma='auto')
SVM.fit(Train_X_Tfidf,Train_Y)
# predict the labels on validation dataset
predictions_SVM = SVM.predict(Test_X_Tfidf)
# Use accuracy_score function to get the accuracy
print("SVM Accuracy Score -> ",accuracy_score(predictions_SVM, Test_Y)*100)
print(classification_report(Test_Y,predictions_SVM))
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from nltk.tokenize import word_tokenize
from nltk import pos_tag
from nltk.corpus import stopwords
from nltk.stem import WordNetLemmatizer
from sklearn.preprocessing import LabelEncoder
from collections import defaultdict
from nltk.corpus import wordnet as wn
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn import model_selection, naive_bayes, svm
from sklearn.metrics import accuracy_score
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
import matplotlib.pyplot as plt
import seaborn as sns
np.random.seed(500)
Corpus = pd.read_csv(r"/amit.csv",encoding='latin-1')
Corpus.head()
Corpus.info()
sns.countplot(Corpus.category)
plt.xlabel('Category')
plt.title('CountPlot')
import nltk
nltk.download('punkt')
nltk.download('wordnet')
nltk.download('averaged_perceptron_tagger')
nltk.download('stopwords')
# 1. Removing Blank Spaces
Corpus['text'].dropna(inplace=True)
# 2. Changing all text to lowercase
Corpus['text_original'] = Corpus['text']
Corpus['text'] = str(Corpus['text'])
Corpus['text'] = [entry.lower() for entry in Corpus['text']]
# 3. Tokenization-In this each entry in the corpus will be broken into set of words
Corpus['text']= [word_tokenize(entry) for entry in Corpus['text']]
# 4. Remove Stop words, Non-Numeric and perfoming Word Stemming/Lemmenting.
# WordNetLemmatizer requires Pos tags to understand if the word is noun or verb or adjective etc. By default it is set to Noun
tag_map = defaultdict(lambda : wn.NOUN)
tag_map['J'] = wn.ADJ
tag_map['V'] = wn.VERB
tag_map['R'] = wn.ADV
Corpus.head()
for index,entry in enumerate(Corpus['text']):
# Declaring Empty List to store the words that follow the rules for this step
Final_words = []
# Initializing WordNetLemmatizer()
word_Lemmatized = WordNetLemmatizer()
# pos_tag function below will provide the 'tag' i.e if the word is Noun(N) or Verb(V) or something else.
for word, tag in pos_tag(entry):
# Below condition is to check for Stop words and consider only alphabets
if word not in stopwords.words('english') and word.isalpha():
word_Final = word_Lemmatized.lemmatize(word,tag_map[tag[0]])
Final_words.append(word_Final)
# The final processed set of words for each iteration will be stored in 'text_final'
Corpus.loc[index,'text_final'] = str(Final_words)
Corpus.drop(['text'], axis=1)
output_path = 'preprocessed_data.csv'
Corpus.to_csv(output_path, index=False)
Train_X, Test_X, Train_Y, Test_Y = model_selection.train_test_split(Corpus['text_final'],Corpus['category'],test_size=0.3)
Encoder = LabelEncoder()
Train_Y = Encoder.fit_transform(Train_Y)
Test_Y = Encoder.fit_transform(Test_Y)
Tfidf_vect = TfidfVectorizer(max_features=5000)
Tfidf_vect.fit(Corpus['text_final'])
Train_X_Tfidf = Tfidf_vect.transform(Train_X)
Test_X_Tfidf = Tfidf_vect.transform(Test_X)
print(Tfidf_vect.vocabulary_)
print(Train_X_Tfidf)
# fit the training dataset on the NB classifier
Naive = naive_bayes.MultinomialNB()
Naive.fit(Train_X_Tfidf,Train_Y)
# predict the labels on validation dataset
predictions_NB = Naive.predict(Test_X_Tfidf)
# Use accuracy_score function to get the accuracy
print("Naive Bayes Accuracy Score -> ",accuracy_score(predictions_NB, Test_Y)*100)
from sklearn import model_selection, naive_bayes, svm
from sklearn.metrics import accuracy_score
Naive = naive_bayes.MultinomialNB()
Naive.fit(Train_X_Tfidf,Train_Y)
# predict the labels on validation dataset
predictions_NB = Naive.predict(Test_X_Tfidf)
# Use accuracy_score function to get the accuracy
print("Naive Bayes Accuracy Score -> ",accuracy_score(predictions_NB, Test_Y)*100)
# Classifier - Algorithm - SVM
# fit the training dataset on the classifier
SVM = svm.SVC(C=1.0, kernel='linear', degree=3, gamma='auto')
SVM.fit(Train_X_Tfidf,Train_Y)
# predict the labels on validation dataset
predictions_SVM = SVM.predict(Test_X_Tfidf)
Train_X_Tfidf.shape
print(classification_report(Test_Y, predictions_NB))
# Classifier - Algorithm - SVM
# fit the training dataset on the classifier
SVM = svm.SVC(C=1.0, kernel='linear', degree=3, gamma='auto')
SVM.fit(Train_X_Tfidf,Train_Y)
# predict the labels on validation dataset
predictions_SVM = SVM.predict(Test_X_Tfidf)
# Use accuracy_score function to get the accuracy
print("SVM Accuracy Score -> ",accuracy_score(predictions_SVM, Test_Y)*100)
print(classification_report(Test_Y,predictions_SVM))
| 0.625324 | 0.783699 |
# Introduction: Finding Prevalence from Observations
In this notebook, we'll look at solving the following statistics problem in a Bayesian Framework. (Problem courtesy of Allen Downey on Twitter: [tweet link](https://twitter.com/AllenDowney/status/1063460263716892674))
> Today's Bayesian problem of the week: Suppose we visit a wild animal preserve where we know that the only animals are lions and tigers and bears, but we don't know how many of each there are.
During the tour, we see 3 lions, 2 tigers, and 1 bear. Assuming that every animal had an equal chance to appear in our sample, estimate the prevalence of each species.
What is the probability that the next animal we see is a bear?
> Solution next week!
Our goal is to estimate the percentage of each species and determine the probability that the next animal is a bear.
## Bayesian Approach
In a frequentist view, we would just use the observed animals to estimate the prevalence and let the data completely speak for itself. In contrast, in a Bayesian framework, we incorporate priors, which here are set to be equal for all species. In the case of infinite observations, the effect of the priors disappear and we only use the data. Due to the limited number of observations, the _priors_ will still have an large effect on the prevalence we obtain, and the _uncertainty_ will be large.
### PyMC3 and MCMC
To solve the problem, we'll build a model in [PyMC3](https://docs.pymc.io/) and then use a variant of Markov Chain Monte Carlo (the No-UTurn Sampler specifically) to draw samples from the posterior. With enough samples, the estimate will converge on the true posterior. Along with maximum likelihood estimates (such as the mean of sampled) values, MCMC also gives us built in uncertainty.
## Model
The overall system is as follows (each part will be explained):
1. The underlying model is a multinomial distribution with parameters $p_k$
2. The _prior_ distribution of $p_k$ is a Dirichlet Distribution
3. The $\alpha$ vector is a parameter of the prior Dirichlet Distribution, hence a _hyperparameter_
4. The prior on $\alpha$ is uniform and is referred to as a _hyperprior_
## Multinomial Distribution
This problem is a classic example of the [multinomial distribution](https://en.wikipedia.org/wiki/Multinomial_distribution) which describes a situation in which we have n independent trials, each with k possible outcomes. With the wildlife preserve problem, n = 6 and k = 3. It is characterized by the probability of each outcome, $p_k$ which must sum to 1. Our goal is to find $p_\text{lions}$, $p_\text{tigers}$, $p_\text{bears}$ given the observations lions: 3, tigers: 2, and bears: 1.
## Dirichlet Distribution
The prior for a multinomial distribution in Bayesian statistics is a [Dirichlet distribution](https://en.wikipedia.org/wiki/Dirichlet_distribution). (Together, a multinomial distribution with a dirichlet prior is called, not surprisingly, a [Dirichlet-Multinomial Distribution](https://en.wikipedia.org/wiki/Dirichlet-multinomial_distribution). The Dirichlet distribution is characterized by $\alpha$, the concentration hyperparameter vector.
### Hyperparameters and Hyperpriors
The $\alpha$ vector is a [hyperparameter](https://en.wikipedia.org/wiki/Hyperparameter), a parameter of a prior distribution. This vector in turn has its _own prior_ distribution which is called a [_hyperprior_](https://en.wikipedia.org/wiki/Hyperprior).
The hyperprior can be thought of as pseudo-counts, which record the number of observations already seen before gathering the data. We want a uniform hyperprior reflecting the chance of observing any species is the same, so we set $\alpha = [1, 1, 1]$.
### Expected Value
One way to get a single point estimate of the prevalence is to use the expected value of the posterior for $p_k$. From Wikipedia, our model is:
$${\begin{array}{lclcl}\mathbf {c} &=&(c_{1},\ldots ,c_{K})&=&{\text{number of occurrences of category }}i=\sum _{j=1}^{N}[x_{j}=i]\\\mathbf {p} \mid \mathbb {X} ,{\boldsymbol {\alpha }}&\sim &\operatorname {Dir} (K,\mathbf {c} +{\boldsymbol {\alpha }})&=&\operatorname {Dir} (K,c_{1}+\alpha _{1},\ldots ,c_{K}+\alpha _{K})\end{array}}$$
which, after filling in the data (number of occurrences) and the hyperpriors becomes:
$${\displaystyle \operatorname {Dir} (K,c_{lions}+\alpha_{lions}, c_{tigers}+\alpha_{tigers}, c_{bears}+\alpha_{bears})}$$
$${\displaystyle \operatorname {Dir} (9, 4, 3, 2)}$$
For a single observation (n = 1), this becomes a [categorical distribution](https://en.wikipedia.org/wiki/Categorical_distribution) and we can get point estimate using the expected value:
$${\displaystyle \operatorname {E} [p_{i}\mid \mathbb {X} ,{\boldsymbol {\alpha }}]={\frac {c_{i}+\alpha _{i}}{N+\sum _{k}\alpha _{k}}}}$$
Therefore, from the viewpoint of a single observation, we get the expected prevalances:
$$p_{lions} = \frac{4}{9} = 44.4\%$$
$$p_{tigers} = \frac{3}{9} = 33.3\%$$
$$p_{bears} = \frac{2}{9} = 22.2\%$$
However, at least to me, this result is unsatisfying because it does not show our uncertainty due to the limited amount of data. For that, we turn to Bayesian modeling and sampling from the posterior with MCMC. For that, we'll move over to PyMC3!
```
import pandas as pd
import numpy as np
# Visualizations
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use('fivethirtyeight')
plt.rcParams['font.size'] = 22
%matplotlib inline
import warnings
warnings.filterwarnings('ignore', category=FutureWarning)
import pymc3 as pm
# Helper functions
from utils import draw_pdf_contours, Dirichlet, plot_points, annotate_plot, add_legend, display_probs
```
# Problem Specifics
```
# observations
animals = ['lions', 'tigers', 'bears']
c = np.array([3, 2, 1])
# hyperparameters (initially all equal)
alphas = np.array([1, 1, 1])
```
# Expected Value
https://en.wikipedia.org/wiki/Categorical_distribution#Bayesian_inference_using_conjugate_prior
$${\displaystyle \operatorname {E} [p_{i}\mid \mathbb {X} ,{\boldsymbol {\alpha }}]={\frac {c_{i}+\alpha _{i}}{N+\sum _{k}\alpha _{k}}}}$$
The expected value is simply as calculated above.
```
display_probs(dict(zip(animals, (alphas + c) / (c.sum() + alphas.sum()))))
display_probs(dict(zip(animals, (4/9, 3/9, 2/9))))
```
# Maximum A Posterior Estimation
The maximum a posterior (MAP) estimate is simply going to be the prevalence seen in the data. This is a frequentist way of viewing the world.
https://en.wikipedia.org/wiki/Categorical_distribution#MAP_estimation
$${\displaystyle \operatorname {arg\,max} \limits _{\mathbf {p} }p(\mathbf {p} \mid \mathbb {X} )={\frac {\alpha _{i}+c_{i}-1}{\sum _{i}(\alpha _{i}+c_{i}-1)}},\qquad \forall i\;\alpha _{i}+c_{i}>1}$$
```
display_probs(dict(zip(animals, (alphas + c - 1) / sum(alphas + c - 1))))
display_probs(dict(zip(animals, c / c.sum())))
```
# Bayesian Model
Now we'll get into building and sampling from a Bayesian model. As a reminder, we are using a multinomial as our model, a Dirichlet distribution as the prior, and uniform _hyperpriors_. The objective is to find the _parameters_ of the prior, $p_k$ which are the probability of each species given the evidence.
$$(\mathbf {p} \mid \mathbb {X}, {\boldsymbol {\alpha}})$$
https://en.wikipedia.org/wiki/Categorical_distribution#Bayesian_inference_using_conjugate_prior
```
with pm.Model() as model:
# Dirichlet hyperparameters are uniform hyperpriors
hyperpriors = pm.Uniform('hyperpriors', shape = 3, observed = alphas)
# Probabilities for each species
parameters = pm.Dirichlet('parameters', a=hyperpriors, shape=3)
# Observed data is a multinomial distribution with 6 trials
observed_data = pm.Multinomial(
'observed_data', n=6, p=parameters, shape=3, observed=c)
model
```
### Sampling from the Model
The cell below samples 1000 draws from the posterior in 2 chains. We use 500 samples for tuning (1000 total because of the 2 chains). This means that for each random variable - the `parameters` - we will have 3000 total samples.
```
with model:
# Sample from the posterior
trace = pm.sample(draws=1000, chains=2, tune=500, discard_tuned_samples=False)
```
## Inspecting Results
```
summary = pm.summary(trace)
summary.index = animals
summary
# Tuning samples
tune_df = pd.DataFrame(trace['parameters'][:1000], columns = animals)
tune_df['tune'] = True
# Samples after tuning
trace_df = pd.DataFrame(trace['parameters'][1000:], columns = animals)
trace_df['tune'] = False
all_df = pd.concat([tune_df, trace_df])
trace_df.head()
```
For a single point estimate, we can use the mean of the samples after burn in.
```
# For probabilities use samples after burn in
pvals = trace_df.iloc[:, :3].mean(axis = 0)
display_probs(dict(zip(animals, pvals)))
```
These numbers align nearly exactly with the expected values! However, we also get a range of uncertainty.
```
trace_df.iloc[:, :3].apply(lambda x: np.percentile(x, 5)).to_frame().rename(columns = {0: '5th percentile'})
trace_df.iloc[:, :3].apply(lambda x: np.percentile(x, 95)).to_frame().rename(columns = {0: '95th percentile'})
```
We can see the large amount of uncertainty in the estimates due to the limited amount of data. To see this visually, we can use some of PyMC3's built in plots.
# Diagnostic Plots
PyMC3 offers a number of [plotting options](https://docs.pymc.io/api/plots.html) for inspecting our samples.
## Posterior Plot
```
ax = pm.plot_posterior(trace, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
```
The 95% HPD is the same idea as a 95% confidence interval in frequentist statistics. In Bayesian stats, this is called the Highest Posterior Density.
## Traceplot
The traceplot shows a kernel density estimate and all the samples that were drawn on the right. We collapse the chains on th plots (`combined = True`) but in reality we drew 2 independent chains.
```
prop_cycle = plt.rcParams['axes.prop_cycle']
cs = [x['color'] for x in list(prop_cycle)]
ax = pm.traceplot(trace, varnames = ['parameters'], figsize = (20, 10), combined = True, skip_first = 1000);
ax[0][0].set_title('KDE of Posteriors'); ax[0][1].set_title('Values in Trace');
add_legend(ax[0][0])
add_legend(ax[0][1])
```
# Maximum A Posteriori Result with PyMC3
```
with model:
# Find the maximum a posteriori estimate
map_ = pm.find_MAP()
display_probs(dict(zip(animals, map_['parameters'])))
```
The MAP estimates are exactly the same as the observations. These are also the results that a frequentist would come up with!
# Sample From Posterior
We can now use the posterior (contained in the `trace`) to draw samples of data. For example, we can simulate 1000 trips to the wildlife preserve as follows.
```
with model:
samples = pm.sample_ppc(trace, samples = 1000)
dict(zip(animals, samples['observed_data'].mean(axis = 0)))
```
These represent the estimate number of each species we'd see over 1000 trips to the preserve.
```
sample_df = pd.DataFrame(samples['observed_data'], columns = animals)
plt.figure(figsize = (22, 8))
for i, animal in enumerate(sample_df):
plt.subplot(1, 3, i+1)
sample_df[animal].value_counts().sort_index().plot.bar(color = 'r');
plt.xticks(range(7), range(7), rotation = 0);
plt.xlabel('Number of Times Seen'); plt.ylabel('Occurences');
plt.title(f'1000 Samples for {animal}');
```
Interestingly, the most likely number of bears is zero!
# Dirichlet Distribution
We can plot the Dirichlet Distribution as a triangle with colors indicating the value of the hyperpriors. This code is taken from the following sources.
http://blog.bogatron.net/blog/2014/02/02/visualizing-dirichlet-distributions/
https://gist.github.com/tboggs/8778945
```
draw_pdf_contours(Dirichlet(alphas))
annotate_plot()
draw_pdf_contours(Dirichlet(6 * pvals))
annotate_plot();
```
# Next Observation
In order to find what we can expect from the next observation, we draw a single sample 10000 from a multinomial distribution. The probability of seeing each species is proportional to that obtained from the sampling.
```
next_obs = np.random.multinomial(n = 1, pvals = pvals, size = 10000)
# Data manipulation
next_obs = pd.melt(pd.DataFrame(next_obs, columns = ['Lions', 'Tigers', 'Bears'])).\
groupby('variable')['value'].\
value_counts(normalize=True).to_frame().\
rename(columns = {'value': 'total'}).reset_index()
next_obs = next_obs.loc[next_obs['value'] == 1]
# Bar plot
next_obs.set_index('variable')['total'].plot.bar(figsize = (8, 6));
plt.title('Next Observation Likelihood');
plt.ylabel('Likelihood'); plt.xlabel('');
next_obs
```
# Tuning vs Broken In Samples
To see if the tuning samples display any major difference from those past the burn-in period, we can plot both distributions. Traditionally, the tuning samples are discarded.
```
all_df = pd.melt(all_df, id_vars=['tune'],
var_name='animal', value_name='posterior')
plt.rcParams['font.size'] = 20
g = sns.FacetGrid(data = all_df, hue = 'tune', col = 'animal', size = 6)
g.map(sns.kdeplot, 'posterior');
l = plt.legend(prop={'size': 20});
l.set_title('Tune');
g.set_ylabels(label = 'Density');
```
# Conclusions
```
summary
```
Given the question posed, we can provide the following answers:
__Estimated Prevalence__
Below are the means and 95% HPD for the estimates
* Lions: 44.5% (16.9% - 75.8%)
* Tigers: 32.7% (6.7% - 60.5%)
* Bears: 22.7% (1.7% - 50.0%)
__Probability Next Observation is a Bear__
Based on the sampling, 22.9%.
(Results may change from one run to the next. This is due to the stochastic nature of MCMC).
The best parts about Bayesian Inference are the incorporation of priors and the uncertainty inherent in the methods. With the scant evidence, we can provide estimates, but only with a large amount of uncertainty. More visits to the wildlife preserve would certainly help to clear up the matter!
## Next Steps
If we have more observations, we can simply add them to the model as follows:
```
c = np.array([[3, 2, 1],
[2, 3, 1],
[3, 2, 1],
[2, 3, 1]])
with pm.Model() as model:
# Dirichlet hyperparameters are uniform hyperpriors
hyperpriors = pm.Uniform('hyperpriors', shape = 3, observed = alphas)
# Probabilities for each species
parameters = pm.Dirichlet('parameters', a=hyperpriors, shape=3)
# Observed data is a multinomial distribution with 6 trials
observed_data = pm.Multinomial(
'observed_data', n=6, p=parameters, shape=3, observed=c)
trace = pm.sample(draws=1000, chains=2, tune=500, discard_tuned_samples=True)
summary = pm.summary(trace)
summary.index = animals
summary
```
The uncertainty of the prevalence of bears has descreased and the prevalence of the lions and tigers is nearly identical as expected. As we gather more data, we can incorporate it into the model to get more accurate estimates.
```
ax = pm.plot_posterior(trace, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
```
## Altering Priors
Below we change the alpha values which places more (higher values) or less (lower values) emphasis on the priors. With a greater emphasis, the posteriors are closer to each other, but with smaller emphasis, the posteriors are more reflective of the observations.
```
# observations
animals = ['lions', 'tigers', 'bears']
c = np.array([3, 2, 1])
alphas = np.array([10, 10, 10])
def sample_with_priors(alphas):
with pm.Model() as model:
# Dirichlet hyperparameters are uniform hyperpriors
# hyperpriors = pm.Uniform('hyperpriors', shape = 3, observed = alphas)
# Probabilities for each species
parameters = pm.Dirichlet('parameters', a=alphas, shape=3)
# Observed data is a multinomial distribution with 6 trials
observed_data = pm.Multinomial(
'observed_data', n=6, p=parameters, shape=3, observed=c)
trace = pm.sample(draws=1000, chains=2, tune=500, discard_tuned_samples=True)
return trace
t = sample_with_priors(np.array([10, 10, 10]))
ax = pm.plot_posterior(t, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
plt.suptitle(f'10 Prior', y = 1.05);
trace_dict = {}
for alpha_array in [np.array([1, 1, 1]), np.array([0.1, 0.1, 0.1]), np.array([0.5, 0.5, 0.5])]:
trace_dict[str(alpha_array[0])] = sample_with_priors(alpha_array)
prior = '0.1'
trace = trace_dict[prior]
ax = pm.plot_posterior(trace, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
plt.suptitle(f'{prior} Prior', y = 1.05);
summary = pm.summary(trace)
summary.index = animals
summary
```
With a smaller weight on the priors, the data has a greater influence on the posterior. Therefore, the mean values are nearly equal to the prevalence in the observations.
```
prior = '0.5'
trace = trace_dict[prior]
ax = pm.plot_posterior(trace, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
plt.suptitle(f'{prior} Prior', y = 1.05);
summary = pm.summary(trace)
summary.index = animals
summary
prior = '1'
trace = trace_dict[prior]
ax = pm.plot_posterior(trace, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
plt.suptitle(f'{prior} Prior', y = 1.05);
summary = pm.summary(trace)
summary.index = animals
summary
```
With the greatest emphasis on the priors, they play a larger role in the posterior estimates and therefore tend to bring the estimated posterior means closer together. The data has a smaller impact when the priors are greater.
Overall, the choice of the hyperpriors depends on our confidence in the prior distribution. If we have good reason to believe all the animals are represented at the same frequency, then we should increase the weight of the priors.
|
github_jupyter
|
import pandas as pd
import numpy as np
# Visualizations
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use('fivethirtyeight')
plt.rcParams['font.size'] = 22
%matplotlib inline
import warnings
warnings.filterwarnings('ignore', category=FutureWarning)
import pymc3 as pm
# Helper functions
from utils import draw_pdf_contours, Dirichlet, plot_points, annotate_plot, add_legend, display_probs
# observations
animals = ['lions', 'tigers', 'bears']
c = np.array([3, 2, 1])
# hyperparameters (initially all equal)
alphas = np.array([1, 1, 1])
display_probs(dict(zip(animals, (alphas + c) / (c.sum() + alphas.sum()))))
display_probs(dict(zip(animals, (4/9, 3/9, 2/9))))
display_probs(dict(zip(animals, (alphas + c - 1) / sum(alphas + c - 1))))
display_probs(dict(zip(animals, c / c.sum())))
with pm.Model() as model:
# Dirichlet hyperparameters are uniform hyperpriors
hyperpriors = pm.Uniform('hyperpriors', shape = 3, observed = alphas)
# Probabilities for each species
parameters = pm.Dirichlet('parameters', a=hyperpriors, shape=3)
# Observed data is a multinomial distribution with 6 trials
observed_data = pm.Multinomial(
'observed_data', n=6, p=parameters, shape=3, observed=c)
model
with model:
# Sample from the posterior
trace = pm.sample(draws=1000, chains=2, tune=500, discard_tuned_samples=False)
summary = pm.summary(trace)
summary.index = animals
summary
# Tuning samples
tune_df = pd.DataFrame(trace['parameters'][:1000], columns = animals)
tune_df['tune'] = True
# Samples after tuning
trace_df = pd.DataFrame(trace['parameters'][1000:], columns = animals)
trace_df['tune'] = False
all_df = pd.concat([tune_df, trace_df])
trace_df.head()
# For probabilities use samples after burn in
pvals = trace_df.iloc[:, :3].mean(axis = 0)
display_probs(dict(zip(animals, pvals)))
trace_df.iloc[:, :3].apply(lambda x: np.percentile(x, 5)).to_frame().rename(columns = {0: '5th percentile'})
trace_df.iloc[:, :3].apply(lambda x: np.percentile(x, 95)).to_frame().rename(columns = {0: '95th percentile'})
ax = pm.plot_posterior(trace, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
prop_cycle = plt.rcParams['axes.prop_cycle']
cs = [x['color'] for x in list(prop_cycle)]
ax = pm.traceplot(trace, varnames = ['parameters'], figsize = (20, 10), combined = True, skip_first = 1000);
ax[0][0].set_title('KDE of Posteriors'); ax[0][1].set_title('Values in Trace');
add_legend(ax[0][0])
add_legend(ax[0][1])
with model:
# Find the maximum a posteriori estimate
map_ = pm.find_MAP()
display_probs(dict(zip(animals, map_['parameters'])))
with model:
samples = pm.sample_ppc(trace, samples = 1000)
dict(zip(animals, samples['observed_data'].mean(axis = 0)))
sample_df = pd.DataFrame(samples['observed_data'], columns = animals)
plt.figure(figsize = (22, 8))
for i, animal in enumerate(sample_df):
plt.subplot(1, 3, i+1)
sample_df[animal].value_counts().sort_index().plot.bar(color = 'r');
plt.xticks(range(7), range(7), rotation = 0);
plt.xlabel('Number of Times Seen'); plt.ylabel('Occurences');
plt.title(f'1000 Samples for {animal}');
draw_pdf_contours(Dirichlet(alphas))
annotate_plot()
draw_pdf_contours(Dirichlet(6 * pvals))
annotate_plot();
next_obs = np.random.multinomial(n = 1, pvals = pvals, size = 10000)
# Data manipulation
next_obs = pd.melt(pd.DataFrame(next_obs, columns = ['Lions', 'Tigers', 'Bears'])).\
groupby('variable')['value'].\
value_counts(normalize=True).to_frame().\
rename(columns = {'value': 'total'}).reset_index()
next_obs = next_obs.loc[next_obs['value'] == 1]
# Bar plot
next_obs.set_index('variable')['total'].plot.bar(figsize = (8, 6));
plt.title('Next Observation Likelihood');
plt.ylabel('Likelihood'); plt.xlabel('');
next_obs
all_df = pd.melt(all_df, id_vars=['tune'],
var_name='animal', value_name='posterior')
plt.rcParams['font.size'] = 20
g = sns.FacetGrid(data = all_df, hue = 'tune', col = 'animal', size = 6)
g.map(sns.kdeplot, 'posterior');
l = plt.legend(prop={'size': 20});
l.set_title('Tune');
g.set_ylabels(label = 'Density');
summary
c = np.array([[3, 2, 1],
[2, 3, 1],
[3, 2, 1],
[2, 3, 1]])
with pm.Model() as model:
# Dirichlet hyperparameters are uniform hyperpriors
hyperpriors = pm.Uniform('hyperpriors', shape = 3, observed = alphas)
# Probabilities for each species
parameters = pm.Dirichlet('parameters', a=hyperpriors, shape=3)
# Observed data is a multinomial distribution with 6 trials
observed_data = pm.Multinomial(
'observed_data', n=6, p=parameters, shape=3, observed=c)
trace = pm.sample(draws=1000, chains=2, tune=500, discard_tuned_samples=True)
summary = pm.summary(trace)
summary.index = animals
summary
ax = pm.plot_posterior(trace, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
# observations
animals = ['lions', 'tigers', 'bears']
c = np.array([3, 2, 1])
alphas = np.array([10, 10, 10])
def sample_with_priors(alphas):
with pm.Model() as model:
# Dirichlet hyperparameters are uniform hyperpriors
# hyperpriors = pm.Uniform('hyperpriors', shape = 3, observed = alphas)
# Probabilities for each species
parameters = pm.Dirichlet('parameters', a=alphas, shape=3)
# Observed data is a multinomial distribution with 6 trials
observed_data = pm.Multinomial(
'observed_data', n=6, p=parameters, shape=3, observed=c)
trace = pm.sample(draws=1000, chains=2, tune=500, discard_tuned_samples=True)
return trace
t = sample_with_priors(np.array([10, 10, 10]))
ax = pm.plot_posterior(t, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
plt.suptitle(f'10 Prior', y = 1.05);
trace_dict = {}
for alpha_array in [np.array([1, 1, 1]), np.array([0.1, 0.1, 0.1]), np.array([0.5, 0.5, 0.5])]:
trace_dict[str(alpha_array[0])] = sample_with_priors(alpha_array)
prior = '0.1'
trace = trace_dict[prior]
ax = pm.plot_posterior(trace, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
plt.suptitle(f'{prior} Prior', y = 1.05);
summary = pm.summary(trace)
summary.index = animals
summary
prior = '0.5'
trace = trace_dict[prior]
ax = pm.plot_posterior(trace, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
plt.suptitle(f'{prior} Prior', y = 1.05);
summary = pm.summary(trace)
summary.index = animals
summary
prior = '1'
trace = trace_dict[prior]
ax = pm.plot_posterior(trace, varnames = ['parameters'],
figsize = (20, 10), edgecolor = 'k');
plt.rcParams['font.size'] = 22
for i, a in enumerate(animals):
ax[i].set_title(a);
plt.suptitle(f'{prior} Prior', y = 1.05);
summary = pm.summary(trace)
summary.index = animals
summary
| 0.721939 | 0.995003 |
# Lecture 11: Dynamic Programming
CBIO (CSCI) 4835/6835: Introduction to Computational Biology
## Overview and Objectives
We've so far discussed sequence alignment from the perspective of distance metrics: Hamming distance and edit distance in particular. However, on the latter point we've been coy; how is edit distance actually computed for arbitrary sequences? How does one decide on the optimal alignment, particularly when using different scoring matrices? We'll go over how all these concepts are incorporated around the concept of *dynamic programming*, and how this allows you to align arbitrary sequences in an optimal way.
By the end of this lecture, you should be able to:
- Describe how dynamic programming works and what its runtime properties are
- Relate dynamic programming to the Manhattan Tourist problem, and why it provides the optimal solution
- Compute the edit distance for two sequences
## Part 1: Change, Revisited
Remember the Change Problem?
Say we want to provide change totaling 97 cents.
Lots of different coin combinations you could use, but if we wanted to use as few coins as possible:
- 3 quarters (75 cents)
- 2 dimes (20 cents)
- 2 pennies (2 cents)
### Two Questions
**1: How do we know this is the fewest possible number of coins?**
**2: Can we generalize to arbitrary denominations (e.g. 3 cent pieces, 9 cent pieces, etc)?**
### Formally
**Problem:** Convert some amount of money $M$ into the given denominations, using the fewest possible number of coins.
**Input:** Amount of money $M$, and an array of $d$ denominations $\vec{c} = (c_1, c_2, ..., c_d)$, sorted in decreasing order (so $c_1 > c_2 > ... > c_d$).
**Output:** A list of $d$ integers, $i_1, i_2, ..., i_d$, such that
$c_1i_1 + c_2i_2 + ... + c_di_d = M$
and
$i_1 + i_2 + ... + i_d$ is as small as possible.
### Yay, Equations...
Let's look at an example.
- **Given:** The denominations $\vec{c} = (1, 3, 5)$ (so $d = 3$)
- **Problem:** What is the minimum number of coins needed to make each of the following values for $M$?

(hopefully, you can see only 1 coin each is needed to make the values $M = 1$, $M = 3$, and $M = 5$)
How about for the other values?

You'll need 2 coins for $M = 2$, $M = 4$, $M = 6$, $M = 8$, and $M = 10$.
**What are the coins?**
See any patterns yet?
How about the remaining values?

3 coins each for $M = 7$ and $M = 9$.
See the pattern yet?
### Recurrence Relations
A *recurrence relation* is, generally speaking, an equation that relies on previous values of the same equation (future values are *functions* of previous values).
What examples of common problems fall under this category?
- Differential Equations
- Fibonacci Numbers
1, 1, 2, 3, 5, 8, 13, 21...
$f(n) = f(n - 1) + f(n - 2)$
So, for our example of having 3 denominations, our recurrence relation looks something like this:

- If $M = 1$, $M = 3$, or $M = 5$, $minNumCoins$ is 0 + 1, so we get 1. These "special cases" are referred to in recurrence relations as **base cases**.
- If $M = 2$, $M = 4$, $M = 6$, or $M = 8$, these all reduce to the base cases, with an added +1, so each of these evaluates to 2.
- Finally, if $M = 7$ or $M = 9$, these are reduced to one of the above cases first (+1), then one of the base cases (+1), for a total of 3.
### So, in general
What would this recurrence relation look like for the *general* case of $d$ denominations?

(see any problems yet?)
### Example, Part Deux
Let's say $M = 77$, and our available denominations are $\vec{c} = (1, 3, 7)$. How does this recurrence relation unfold?
Well...





Notice how many times that "70" appeared?
The reason it was highlighted in a red circle is to draw attention to the fact that it's being **re-computed at every one of those steps.**
### So many repeated calculations!
At multiple levels of the recurrence tree, it's redoing the same calculations over and over.
- In our example of $M = 77$, $\vec{c} = (1, 3, 7)$, the optimal coin combination for 70 is computed **9 separate times**.
- The optimal coin combination for 50 cents is computed **billions of times**.
- How about the optimal coin combination for 3 cents? o_O
**How can we improve the algorithm so we don't waste so much time recomputing the same values over and over?**
## Part 2: Dynamic Programming
The idea is pretty simple, actually: instead of re-computing values in our algorithm, let's **save the results of each computation for all amounts 0 to $M$.**
Therefore, we can just "look up" the answer for a value that's already been computed.
This new approach should give us a runtime complexity of $\mathcal{O}(Md)$, where $M$ is the amount of money and $d$ is the number of denominations (what was the runtime before?).
This is called **dynamic programming.**
Let's look at a modification of the example from before, with $M = 9$ and $\vec{c} = (1, 3, 7)$.


If that looked and felt a lot like what we were doing before, that's not wrong!
Dynamic Programming does indeed closely resemble the recurrence relation it is intended to replace.
The difference is, with the recurrence, we had to constantly recompute the "easier" values farther down the tree, since we always started from the top.
With dynamic programming, it's the other way around: we start at the bottom with the "easier" values, and build up to the more complex ones, using the solutions we obtain along the way. In doing so, we avoid repetition.
## Part 3: The Tourist in Manhattan
Imagine you're a tourist in Manhattan.
You're about to leave town (starting at the hotel), but on your way to the subway station, you want to see as many attractions as possible (marked by the red stars).
Your time is limited--you can only move South or East. **What's the "best" path through town?** (meaning the one with the most attractions)


### Formally
Yes, the [Manhattan Tourist Problem](https://en.wikipedia.org/wiki/Travelling_salesman_problem) is indeed a formal problem from Computer Science, and specifically graph theory:
**Problem:** Find the optimal path in a [weighted] grid.
**Input:** A weighted grid $G$ with two labeled vertices: a *source* (the starting point) and a *sink* (the ending point).
**Output:** The optimal path through $G$, starting at the source and ending at the sink.
### First attempt: the "greedy" approach
One reasonable first attempt, as I would have with the Change problem, would be a *greedy* approach: **every time I have to make a decision, pick the best one available**.
With the Manhattan Tourist Problem, this means that at each intersection, choose the direction (south or east) that gives me immediate access to the most attractions.









What's wrong with this approach?
It can miss the *global* optimum, if it chooses a route early on that diverts it away:

This is the optimal route, with a total weight of 22. However, what route would a greedy approach choose?

The red route has only a global weight of 18, but the initial choice at the source--between 5 and 1--will push the greedy algorithm off course.
### Dynamic Programming to the Rescue
Hopefully by now you're already thinking "this sounds like something dynamic programming could help with."
- At each vertex (intersection) in the graph, we calculate the optimal score to get there
- A given vertex's score is the maximum of the incoming edge weights + the previous vertex's score (sound familiar?)

The gold edges represent those which the algorithm selects as "optimal" for each vertex.






Once we've reached the sink, it's a simple matter of backtracking along the gold edges to find the optimal route (which we highlight in green here).
### Complexity
With the change problem, we said the runtime complexity of dynamic programming was $\mathcal{O}(Md)$, where $M$ is the amount of money, and $d$ is the number of denominations.
Let's make this a bit more formal. We have a graph / matrix, and each intersection $s_{i,j}$ has a score according to the recurrence:

For a matrix with $n$ rows and $m$ columns, what is the complexity?
$\mathcal{O}(nm)$. Basically, we have to look at every element of the matrix.
But that's still better than the recurrence relation we saw earlier!
## Part 4: Sequence Alignment
So how does all this relate to sequence alignment? How does dynamic programming play into finding the longest common subsequence of two polypeptides or nucleic acid sequences?
Given two sequences, let's use dynamic programming to find their best alignment.
$v$: `ATCTGATC`
$w$: `TGCATAC`
Our nucleotide string $v$ has length 8, and $w$ has length 7. How can we align these two sequences optimally?
### Alignment Matrix
We can represent these strings along the rows and columns of an *alignment matrix.*
Assign one sequence to the rows, and one sequence to the columns.

At each intersection / vertex, we have three options:
- Go south (insertion / deletion)
- Go east (deletion / deletion)
- Go south-east (match / mismatch)
Every diagonal movement represents a match. We can immediately see all our common subsequences this way:

Now, we just need to join up as many of these aligned subsequences as possible to make the **longest common subsequence**, and hence, the optimal alignment.
The full path, from source (upper left) to sink (bottom right), represents a common sequence.

### Using the Alignment Matrix for Edit Distance
- Every alignment of two sequences corresponds to a path in the alignment matrix from source to sink
- Horizontal and vertical edges correspond to indels (insertions and deletions)
- Diagonal edges correspond to matches and mismatches
### Dynamic Programming for Sequence Alignment
Let's see how this would play out algorithmically. Here's an example:
$v$ = `ATCGTAC`
$w$ = `ATGTTAC`
One possible alignment of the two sequences might look like this ($v$ on top, $w$ on bottom)

So the corresponding alignment matrix would have a path from source to sink like this:

Programmatically, it would follow these steps.
**Step 1:** Initialize the $0^{th}$ row and $0^{th}$ column to be all 0s.

**Step 2:** Use the following recurrence formula to calculate $s_{i,j}$ for each $i$ and $j$ in the matrix:

- Top: a match (or mismatch)
- Middle: a deletion (with respect to $v$)
- Bottom: an insertion (with respect to $v$)
**You'll pretty much run Step 2 over and over until the matrix is filled.**

You'll look for any matches first (in red), then fill in the indels.







We've filled the alignment matrix! Now how do we assemble the final, optimal alignment of the two sequences?
Start at the sink and follow the arrows back!

### Pseudocode

### Some final thoughts on Dynamic Programming
- How could we set up this problem in Python? What would the data structures be?
- Remember edit distance? It's the measure of how different two sequences are. By contrast, the alignment score from dynamic programming is a _similarity score_. If edit distance is 0, what do we expect the alignment score to be?
- How could we modify the scoring procedure in dynamic programming to allow for scoring matrices like PAM and BLOSUM?
## Administrivia
- Grades for Assignments 1 and 2 are on eLC! Ping me with questions.
- If you're struggling with the assignments, let's meet and go over these questions.
- Assignment 3 is out, and will be the last assignment before the "midterm".
- Regarding the midterm--does everyone have a laptop they could bring to class for the exam?
## Additional Resources
1. Compeau, Phillip. *An Introduction to Bioinformatics*. Dynamic Programming: Edit Distance, [Part 1](http://compeau.cbd.cmu.edu/teaching/jones-pevzner-slides/edit-distance-part-1/) and [Part 2](http://compeau.cbd.cmu.edu/teaching/jones-pevzner-slides/edit-distance-part-2/).
|
github_jupyter
|
# Lecture 11: Dynamic Programming
CBIO (CSCI) 4835/6835: Introduction to Computational Biology
## Overview and Objectives
We've so far discussed sequence alignment from the perspective of distance metrics: Hamming distance and edit distance in particular. However, on the latter point we've been coy; how is edit distance actually computed for arbitrary sequences? How does one decide on the optimal alignment, particularly when using different scoring matrices? We'll go over how all these concepts are incorporated around the concept of *dynamic programming*, and how this allows you to align arbitrary sequences in an optimal way.
By the end of this lecture, you should be able to:
- Describe how dynamic programming works and what its runtime properties are
- Relate dynamic programming to the Manhattan Tourist problem, and why it provides the optimal solution
- Compute the edit distance for two sequences
## Part 1: Change, Revisited
Remember the Change Problem?
Say we want to provide change totaling 97 cents.
Lots of different coin combinations you could use, but if we wanted to use as few coins as possible:
- 3 quarters (75 cents)
- 2 dimes (20 cents)
- 2 pennies (2 cents)
### Two Questions
**1: How do we know this is the fewest possible number of coins?**
**2: Can we generalize to arbitrary denominations (e.g. 3 cent pieces, 9 cent pieces, etc)?**
### Formally
**Problem:** Convert some amount of money $M$ into the given denominations, using the fewest possible number of coins.
**Input:** Amount of money $M$, and an array of $d$ denominations $\vec{c} = (c_1, c_2, ..., c_d)$, sorted in decreasing order (so $c_1 > c_2 > ... > c_d$).
**Output:** A list of $d$ integers, $i_1, i_2, ..., i_d$, such that
$c_1i_1 + c_2i_2 + ... + c_di_d = M$
and
$i_1 + i_2 + ... + i_d$ is as small as possible.
### Yay, Equations...
Let's look at an example.
- **Given:** The denominations $\vec{c} = (1, 3, 5)$ (so $d = 3$)
- **Problem:** What is the minimum number of coins needed to make each of the following values for $M$?

(hopefully, you can see only 1 coin each is needed to make the values $M = 1$, $M = 3$, and $M = 5$)
How about for the other values?

You'll need 2 coins for $M = 2$, $M = 4$, $M = 6$, $M = 8$, and $M = 10$.
**What are the coins?**
See any patterns yet?
How about the remaining values?

3 coins each for $M = 7$ and $M = 9$.
See the pattern yet?
### Recurrence Relations
A *recurrence relation* is, generally speaking, an equation that relies on previous values of the same equation (future values are *functions* of previous values).
What examples of common problems fall under this category?
- Differential Equations
- Fibonacci Numbers
1, 1, 2, 3, 5, 8, 13, 21...
$f(n) = f(n - 1) + f(n - 2)$
So, for our example of having 3 denominations, our recurrence relation looks something like this:

- If $M = 1$, $M = 3$, or $M = 5$, $minNumCoins$ is 0 + 1, so we get 1. These "special cases" are referred to in recurrence relations as **base cases**.
- If $M = 2$, $M = 4$, $M = 6$, or $M = 8$, these all reduce to the base cases, with an added +1, so each of these evaluates to 2.
- Finally, if $M = 7$ or $M = 9$, these are reduced to one of the above cases first (+1), then one of the base cases (+1), for a total of 3.
### So, in general
What would this recurrence relation look like for the *general* case of $d$ denominations?

(see any problems yet?)
### Example, Part Deux
Let's say $M = 77$, and our available denominations are $\vec{c} = (1, 3, 7)$. How does this recurrence relation unfold?
Well...





Notice how many times that "70" appeared?
The reason it was highlighted in a red circle is to draw attention to the fact that it's being **re-computed at every one of those steps.**
### So many repeated calculations!
At multiple levels of the recurrence tree, it's redoing the same calculations over and over.
- In our example of $M = 77$, $\vec{c} = (1, 3, 7)$, the optimal coin combination for 70 is computed **9 separate times**.
- The optimal coin combination for 50 cents is computed **billions of times**.
- How about the optimal coin combination for 3 cents? o_O
**How can we improve the algorithm so we don't waste so much time recomputing the same values over and over?**
## Part 2: Dynamic Programming
The idea is pretty simple, actually: instead of re-computing values in our algorithm, let's **save the results of each computation for all amounts 0 to $M$.**
Therefore, we can just "look up" the answer for a value that's already been computed.
This new approach should give us a runtime complexity of $\mathcal{O}(Md)$, where $M$ is the amount of money and $d$ is the number of denominations (what was the runtime before?).
This is called **dynamic programming.**
Let's look at a modification of the example from before, with $M = 9$ and $\vec{c} = (1, 3, 7)$.


If that looked and felt a lot like what we were doing before, that's not wrong!
Dynamic Programming does indeed closely resemble the recurrence relation it is intended to replace.
The difference is, with the recurrence, we had to constantly recompute the "easier" values farther down the tree, since we always started from the top.
With dynamic programming, it's the other way around: we start at the bottom with the "easier" values, and build up to the more complex ones, using the solutions we obtain along the way. In doing so, we avoid repetition.
## Part 3: The Tourist in Manhattan
Imagine you're a tourist in Manhattan.
You're about to leave town (starting at the hotel), but on your way to the subway station, you want to see as many attractions as possible (marked by the red stars).
Your time is limited--you can only move South or East. **What's the "best" path through town?** (meaning the one with the most attractions)


### Formally
Yes, the [Manhattan Tourist Problem](https://en.wikipedia.org/wiki/Travelling_salesman_problem) is indeed a formal problem from Computer Science, and specifically graph theory:
**Problem:** Find the optimal path in a [weighted] grid.
**Input:** A weighted grid $G$ with two labeled vertices: a *source* (the starting point) and a *sink* (the ending point).
**Output:** The optimal path through $G$, starting at the source and ending at the sink.
### First attempt: the "greedy" approach
One reasonable first attempt, as I would have with the Change problem, would be a *greedy* approach: **every time I have to make a decision, pick the best one available**.
With the Manhattan Tourist Problem, this means that at each intersection, choose the direction (south or east) that gives me immediate access to the most attractions.









What's wrong with this approach?
It can miss the *global* optimum, if it chooses a route early on that diverts it away:

This is the optimal route, with a total weight of 22. However, what route would a greedy approach choose?

The red route has only a global weight of 18, but the initial choice at the source--between 5 and 1--will push the greedy algorithm off course.
### Dynamic Programming to the Rescue
Hopefully by now you're already thinking "this sounds like something dynamic programming could help with."
- At each vertex (intersection) in the graph, we calculate the optimal score to get there
- A given vertex's score is the maximum of the incoming edge weights + the previous vertex's score (sound familiar?)

The gold edges represent those which the algorithm selects as "optimal" for each vertex.






Once we've reached the sink, it's a simple matter of backtracking along the gold edges to find the optimal route (which we highlight in green here).
### Complexity
With the change problem, we said the runtime complexity of dynamic programming was $\mathcal{O}(Md)$, where $M$ is the amount of money, and $d$ is the number of denominations.
Let's make this a bit more formal. We have a graph / matrix, and each intersection $s_{i,j}$ has a score according to the recurrence:

For a matrix with $n$ rows and $m$ columns, what is the complexity?
$\mathcal{O}(nm)$. Basically, we have to look at every element of the matrix.
But that's still better than the recurrence relation we saw earlier!
## Part 4: Sequence Alignment
So how does all this relate to sequence alignment? How does dynamic programming play into finding the longest common subsequence of two polypeptides or nucleic acid sequences?
Given two sequences, let's use dynamic programming to find their best alignment.
$v$: `ATCTGATC`
$w$: `TGCATAC`
Our nucleotide string $v$ has length 8, and $w$ has length 7. How can we align these two sequences optimally?
### Alignment Matrix
We can represent these strings along the rows and columns of an *alignment matrix.*
Assign one sequence to the rows, and one sequence to the columns.

At each intersection / vertex, we have three options:
- Go south (insertion / deletion)
- Go east (deletion / deletion)
- Go south-east (match / mismatch)
Every diagonal movement represents a match. We can immediately see all our common subsequences this way:

Now, we just need to join up as many of these aligned subsequences as possible to make the **longest common subsequence**, and hence, the optimal alignment.
The full path, from source (upper left) to sink (bottom right), represents a common sequence.

### Using the Alignment Matrix for Edit Distance
- Every alignment of two sequences corresponds to a path in the alignment matrix from source to sink
- Horizontal and vertical edges correspond to indels (insertions and deletions)
- Diagonal edges correspond to matches and mismatches
### Dynamic Programming for Sequence Alignment
Let's see how this would play out algorithmically. Here's an example:
$v$ = `ATCGTAC`
$w$ = `ATGTTAC`
One possible alignment of the two sequences might look like this ($v$ on top, $w$ on bottom)

So the corresponding alignment matrix would have a path from source to sink like this:

Programmatically, it would follow these steps.
**Step 1:** Initialize the $0^{th}$ row and $0^{th}$ column to be all 0s.

**Step 2:** Use the following recurrence formula to calculate $s_{i,j}$ for each $i$ and $j$ in the matrix:

- Top: a match (or mismatch)
- Middle: a deletion (with respect to $v$)
- Bottom: an insertion (with respect to $v$)
**You'll pretty much run Step 2 over and over until the matrix is filled.**

You'll look for any matches first (in red), then fill in the indels.







We've filled the alignment matrix! Now how do we assemble the final, optimal alignment of the two sequences?
Start at the sink and follow the arrows back!

### Pseudocode

### Some final thoughts on Dynamic Programming
- How could we set up this problem in Python? What would the data structures be?
- Remember edit distance? It's the measure of how different two sequences are. By contrast, the alignment score from dynamic programming is a _similarity score_. If edit distance is 0, what do we expect the alignment score to be?
- How could we modify the scoring procedure in dynamic programming to allow for scoring matrices like PAM and BLOSUM?
## Administrivia
- Grades for Assignments 1 and 2 are on eLC! Ping me with questions.
- If you're struggling with the assignments, let's meet and go over these questions.
- Assignment 3 is out, and will be the last assignment before the "midterm".
- Regarding the midterm--does everyone have a laptop they could bring to class for the exam?
## Additional Resources
1. Compeau, Phillip. *An Introduction to Bioinformatics*. Dynamic Programming: Edit Distance, [Part 1](http://compeau.cbd.cmu.edu/teaching/jones-pevzner-slides/edit-distance-part-1/) and [Part 2](http://compeau.cbd.cmu.edu/teaching/jones-pevzner-slides/edit-distance-part-2/).
| 0.942255 | 0.974483 |
ERROR: type should be string, got "https://arxiv.org/pdf/1810.04805.pdf\n\n```\nimport os\nos.sys.path.append('..')\n%matplotlib inline\n%reload_ext autoreload\n%autoreload 2\nfrom __future__ import absolute_import\nfrom __future__ import division\nfrom __future__ import print_function\n\nimport argparse\nimport collections\nimport logging\nimport json\nimport math\nimport os\nimport random\nimport six\nfrom tqdm import tqdm_notebook as tqdm\nfrom IPython.display import HTML, display\n\nimport numpy as np\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torch.utils.data import TensorDataset, DataLoader, RandomSampler, SequentialSampler\nfrom torch.utils.data.distributed import DistributedSampler\n\nimport tokenization\nfrom modeling import BertConfig, BertForMaskedLanguageModelling\nfrom optimization import BERTAdam\nfrom masked_language_model import notqdm, convert_tokens_to_features, LMProcessor, predict_masked_words, predict_next_words\n\nlogging.basicConfig(format = '%(asctime)s - %(levelname)s - %(name)s - %(message)s', \n datefmt = '%m/%d/%Y %H:%M:%S',\n level = logging.INFO)\nlogger = logging.getLogger(__name__)\n```\n\n# Args\n\n```\nparser = argparse.ArgumentParser()\n\n## Required parameters\nparser.add_argument(\"--data_dir\",\n default=None,\n type=str,\n required=True,\n help=\"The input data dir. Should contain the .tsv files (or other data files) for the task.\")\nparser.add_argument(\"--bert_config_file\",\n default=None,\n type=str,\n required=True,\n help=\"The config json file corresponding to the pre-trained BERT model. \\n\"\n \"This specifies the model architecture.\")\nparser.add_argument(\"--task_name\",\n default=None,\n type=str,\n required=True,\n help=\"The name of the task to train.\")\nparser.add_argument(\"--vocab_file\",\n default=None,\n type=str,\n required=True,\n help=\"The vocabulary file that the BERT model was trained on.\")\nparser.add_argument(\"--output_dir\",\n default=None,\n type=str,\n required=True,\n help=\"The output directory where the model checkpoints will be written.\")\n\n## Other parameters\nparser.add_argument(\"--init_checkpoint\",\n default=None,\n type=str,\n help=\"Initial checkpoint (usually from a pre-trained BERT model).\")\nparser.add_argument(\"--do_lower_case\",\n default=False,\n action='store_true',\n help=\"Whether to lower case the input text. True for uncased models, False for cased models.\")\nparser.add_argument(\"--max_seq_length\",\n default=128,\n type=int,\n help=\"The maximum total input sequence length after WordPiece tokenization. \\n\"\n \"Sequences longer than this will be truncated, and sequences shorter \\n\"\n \"than this will be padded.\")\nparser.add_argument(\"--do_train\",\n default=False,\n action='store_true',\n help=\"Whether to run training.\")\nparser.add_argument(\"--do_eval\",\n default=False,\n action='store_true',\n help=\"Whether to run eval on the dev set.\")\nparser.add_argument(\"--train_batch_size\",\n default=32,\n type=int,\n help=\"Total batch size for training.\")\nparser.add_argument(\"--eval_batch_size\",\n default=8,\n type=int,\n help=\"Total batch size for eval.\")\nparser.add_argument(\"--learning_rate\",\n default=5e-5,\n type=float,\n help=\"The initial learning rate for Adam.\")\nparser.add_argument(\"--num_train_epochs\",\n default=3.0,\n type=float,\n help=\"Total number of training epochs to perform.\")\nparser.add_argument(\"--warmup_proportion\",\n default=0.1,\n type=float,\n help=\"Proportion of training to perform linear learning rate warmup for. \"\n \"E.g., 0.1 = 10%% of training.\")\nparser.add_argument(\"--no_cuda\",\n default=False,\n action='store_true',\n help=\"Whether not to use CUDA when available\")\nparser.add_argument(\"--local_rank\",\n type=int,\n default=-1,\n help=\"local_rank for distributed training on gpus\")\nparser.add_argument('--seed', \n type=int, \n default=42,\n help=\"random seed for initialization\")\nparser.add_argument('--gradient_accumulation_steps',\n type=int,\n default=1,\n help=\"Number of updates steps to accumualte before performing a backward/update pass.\")\nexperiment_name = 'horror_uncased_5_tied_mlm'\n\nargv = \"\"\"\n--task_name lm \\\n--data_dir {DATA_DIR} \\\n--vocab_file {BERT_BASE_DIR}/vocab.txt \\\n--bert_config_file {BERT_BASE_DIR}/bert_config.json \\\n--init_checkpoint {BERT_BASE_DIR}/pytorch_model.bin \\\n--do_train \\\n--do_eval \\\n--gradient_accumulation_steps 2 \\\n--train_batch_size 32 \\\n--learning_rate 3e-5 \\\n--num_train_epochs 3.0 \\\n--max_seq_length 128 \\\n--output_dir ../outputs/{name}/\n\"\"\".format(\n BERT_BASE_DIR='../data/weights/cased_L-12_H-768_A-12',\n DATA_DIR='../data/input/horror_gutenberg',\n name=experiment_name\n).replace('\\n', '').split(' ')\nprint(argv)\nargs = parser.parse_args(argv)\n```\n\n# Init\n\n```\nif args.local_rank == -1 or args.no_cuda:\n device = torch.device(\"cuda\" if torch.cuda.is_available() and not args.no_cuda else \"cpu\")\n n_gpu = torch.cuda.device_count()\nelse:\n device = torch.device(\"cuda\", args.local_rank)\n n_gpu = 1\n # Initializes the distributed backend which will take care of sychronizing nodes/GPUs\n torch.distributed.init_process_group(backend='nccl')\nlogger.info(\"device %s n_gpu %d distributed training %r\", device, n_gpu, bool(args.local_rank != -1))\n\nif args.gradient_accumulation_steps < 1:\n raise ValueError(\"Invalid gradient_accumulation_steps parameter: {}, should be >= 1\".format(\n args.gradient_accumulation_steps))\n\nargs.train_batch_size = int(args.train_batch_size / args.gradient_accumulation_steps)\n\nrandom.seed(args.seed)\nnp.random.seed(args.seed)\ntorch.manual_seed(args.seed)\nif n_gpu > 0:\n torch.cuda.manual_seed_all(args.seed)\n\nif not args.do_train and not args.do_eval:\n raise ValueError(\"At least one of `do_train` or `do_eval` must be True.\")\nbert_config = BertConfig.from_json_file(args.bert_config_file)\n\nif args.max_seq_length > bert_config.max_position_embeddings:\n raise ValueError(\n \"Cannot use sequence length {} because the BERT model was only trained up to sequence length {}\".format(\n args.max_seq_length, bert_config.max_position_embeddings))\nif os.path.exists(args.output_dir) and os.listdir(args.output_dir):\n print(\"Output directory ({}) already exists and is not empty.\".format(args.output_dir))\nos.makedirs(args.output_dir, exist_ok=True)\nsave_path = os.path.join(args.output_dir, 'state_dict.pkl')\nsave_path\n```\n\n# Load data\n\n```\ntokenizer = tokenization.FullTokenizer(\n vocab_file=args.vocab_file, do_lower_case=args.do_lower_case)\n\ndecoder = {v:k for k,v in tokenizer.wordpiece_tokenizer.vocab.items()}\nprocessors = {\n \"lm\": LMProcessor,\n}\n \ntask_name = args.task_name.lower()\nif task_name not in processors:\n raise ValueError(\"Task not found: %s\" % (task_name))\n\nprocessor = processors[task_name](tokenizer=tokenizer)\nlabel_list = processor.get_labels()\ntrain_examples = processor.get_train_examples(args.data_dir, skip=30, tqdm=tqdm)\nnum_train_steps = int(\n len(train_examples) / args.train_batch_size * args.num_train_epochs)\ntrain_features = convert_tokens_to_features(\n train_examples, label_list, args.max_seq_length, tokenizer, tqdm=tqdm)\n\nall_input_ids = torch.tensor([f.input_ids for f in train_features], dtype=torch.long)\nall_input_mask = torch.tensor([f.input_mask for f in train_features], dtype=torch.long)\nall_segment_ids = torch.tensor([f.segment_ids for f in train_features], dtype=torch.long)\nall_label_ids = torch.tensor([f.label_id for f in train_features], dtype=torch.long)\nall_label_weights = torch.tensor([f.label_weights for f in train_features], dtype=torch.long)\ntrain_data = TensorDataset(all_input_ids, all_input_mask, all_segment_ids, all_label_ids, all_label_weights)\nif args.local_rank == -1:\n train_sampler = RandomSampler(train_data)\nelse:\n train_sampler = DistributedSampler(train_data)\ntrain_dataloader = DataLoader(train_data, sampler=train_sampler, batch_size=args.train_batch_size)\n```\n\n# Load model\n\n```\nmodel = BertForMaskedLanguageModelling(bert_config)\nif args.init_checkpoint is not None:\n model.bert.load_state_dict(torch.load(args.init_checkpoint, map_location='cpu'))\n \nif os.path.isfile(save_path):\n model.load_state_dict(torch.load(save_path, map_location='cpu'))\n \nmodel.to(device)\n\nif args.local_rank != -1:\n model = torch.nn.parallel.DistributedDataParallel(model, device_ids=[args.local_rank],\n output_device=args.local_rank)\nelif n_gpu > 1:\n model = torch.nn.DataParallel(model)\n \nmodel\n```\n\n# Opt\n\n```\nno_decay = ['bias', 'gamma', 'beta']\noptimizer_parameters = [\n {'params': [p for n, p in model.named_parameters() if n not in no_decay], 'weight_decay_rate': 0.01},\n {'params': [p for n, p in model.named_parameters() if n in no_decay], 'weight_decay_rate': 0.0}\n ]\n\noptimizer = BERTAdam(optimizer_parameters,\n lr=args.learning_rate,\n warmup=args.warmup_proportion,\n t_total=num_train_steps)\n```\n\n# Train 3\n\n```\nval_test=\"\"\"Another gentleman has fallen a victim to the terrible epidemic of suicide which for the last month has prevailed in the West End. Mr. Sidney Crashaw, of Stoke House, Fulham, and King's Pomeroy, Devon, was found, after a prolonged search, hanging dead from the branch of a tree in his garden at one o'clock today. The deceased gentleman dined last night at the Carlton Club and seemed in his usual health and spirits. He left the club at about ten o'clock, and was seen walking leisurely up St. James's Street a little later. Subsequent to this his movements cannot be traced\"\"\"\ndisplay(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=30, T=1, device=device))\ndisplay(predict_masked_words(val_test, processor, tokenizer, model, device=device, max_seq_length=args.max_seq_length))\nglobal_step = 0\n1/0\nmodel.train()\nfor _ in tqdm(range(int(args.num_train_epochs)), desc=\"Epoch\"):\n tr_loss, nb_tr_examples, nb_tr_steps = 0, 0, 0\n with tqdm(total=len(train_dataloader), desc='Iteration', mininterval=0.5) as prog:\n for step, batch in enumerate(train_dataloader):\n batch = tuple(t.to(device) for t in batch)\n input_ids, input_mask, segment_ids, label_ids, label_weights = batch\n \n loss, logits = model(input_ids, segment_ids, input_mask, label_ids, label_weights)\n if n_gpu > 1:\n loss = loss.mean() # mean() to average on multi-gpu.\n if args.gradient_accumulation_steps > 1:\n loss = loss / args.gradient_accumulation_steps\n loss.backward()\n tr_loss += loss.item()\n nb_tr_examples += input_ids.size(0)\n nb_tr_steps += 1\n if (step + 1) % args.gradient_accumulation_steps == 0:\n optimizer.step() # We have accumulated enougth gradients\n model.zero_grad()\n prog.update(1)\n prog.desc = 'Iter. loss={:2.6f}'.format(tr_loss/nb_tr_examples)\n if step%3000==10:\n \n print('step', step, 'loss', tr_loss/nb_tr_examples)\n display(predict_masked_words(val_test, processor, tokenizer, model, device=device, max_seq_length=args.max_seq_length))\n display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=10, device=device))\n tr_loss, nb_tr_examples, nb_tr_steps = 0, 0, 0\n \n \n torch.save(model.state_dict(), save_path)\n\nglobal_step += 1\ntorch.save(model.state_dict(), save_path)\n# val_test=\"\"\"Another gentleman has fallen a victim to the terrible epidemic of suicide which for the last month has prevailed in the West End. Mr. Sidney Crashaw, of Stoke House, Fulham, and King's Pomeroy, Devon, was found, after a prolonged search, hanging dead from the branch of a tree in his garden at one o'clock today. The deceased gentleman dined last night at the Carlton Club and seemed in his usual health and spirits. He left the club at about ten o'clock, and was seen walking leisurely up St. James's Street a little later. Subsequent to this his movements cannot be traced. On the discovery of the body medical aid was at once summoned, but life had evidently been long extinct. So far as is known, Mr. Crashaw had no trouble or anxiety of any kind. This painful suicide, it will be remembered, is the fifth of the kind in the last month. The authorities at Scotland Yard are unable to suggest any explanation of these terrible occurrences.\"\"\"\n# display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=100, T=.5, device=device))\nval_test=\"\"\"His mind spun in on itself . . . .\"\"\"\ndisplay(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=150, T=.2, device=device))\nval_test=\"\"\"A giant spider descended on to . . . .\"\"\"\ndisplay(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=100, T=.5, device=device))\nval_test=\"\"\"Madness enveloped his mind as . . . .\"\"\"\ndisplay(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=100, T=.5, device=device))\nval_test=\"\"\"A thin film of . . . .\"\"\"\ndisplay(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=100, T=.01, device=device))\nval_test=\"\"\"Quivering with fear, he trembled as . . . .\"\"\"\ndisplay(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=100, T=.7, device=device))\nval_test=\"\"\"Madness enveloped his mind as . . . .\"\"\"\ndisplay(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=128, T=.1, device=device))\nval_test=\"\"\"All at once, in a moment of realisation, he knew the secret to creating true artificial intelligence was . . . .\"\"\"\ndisplay(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=300, T=.5, device=device))\nimprove_words_recursive(text, processor, tokenizer, model, iterations=100, max_seq_length=128, n=10, T=0.3, device=\"cuda\", debug=10)\n```\n\n# TODO\n\n- show probability in next word logging. Record probability of each letter, then use them when displaying as html\n- try other ways of doing next word. E.g. going back and redoing, doing more than 1 at once\n- make the masked language generator often mask last word\n- should I be doing loss on just the masked words, or all? It's hard to tell from the tensorflow repo. This is marked with a TODO or FIXME in the code\n\n" |
github_jupyter
|
import os
os.sys.path.append('..')
%matplotlib inline
%reload_ext autoreload
%autoreload 2
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import argparse
import collections
import logging
import json
import math
import os
import random
import six
from tqdm import tqdm_notebook as tqdm
from IPython.display import HTML, display
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import TensorDataset, DataLoader, RandomSampler, SequentialSampler
from torch.utils.data.distributed import DistributedSampler
import tokenization
from modeling import BertConfig, BertForMaskedLanguageModelling
from optimization import BERTAdam
from masked_language_model import notqdm, convert_tokens_to_features, LMProcessor, predict_masked_words, predict_next_words
logging.basicConfig(format = '%(asctime)s - %(levelname)s - %(name)s - %(message)s',
datefmt = '%m/%d/%Y %H:%M:%S',
level = logging.INFO)
logger = logging.getLogger(__name__)
parser = argparse.ArgumentParser()
## Required parameters
parser.add_argument("--data_dir",
default=None,
type=str,
required=True,
help="The input data dir. Should contain the .tsv files (or other data files) for the task.")
parser.add_argument("--bert_config_file",
default=None,
type=str,
required=True,
help="The config json file corresponding to the pre-trained BERT model. \n"
"This specifies the model architecture.")
parser.add_argument("--task_name",
default=None,
type=str,
required=True,
help="The name of the task to train.")
parser.add_argument("--vocab_file",
default=None,
type=str,
required=True,
help="The vocabulary file that the BERT model was trained on.")
parser.add_argument("--output_dir",
default=None,
type=str,
required=True,
help="The output directory where the model checkpoints will be written.")
## Other parameters
parser.add_argument("--init_checkpoint",
default=None,
type=str,
help="Initial checkpoint (usually from a pre-trained BERT model).")
parser.add_argument("--do_lower_case",
default=False,
action='store_true',
help="Whether to lower case the input text. True for uncased models, False for cased models.")
parser.add_argument("--max_seq_length",
default=128,
type=int,
help="The maximum total input sequence length after WordPiece tokenization. \n"
"Sequences longer than this will be truncated, and sequences shorter \n"
"than this will be padded.")
parser.add_argument("--do_train",
default=False,
action='store_true',
help="Whether to run training.")
parser.add_argument("--do_eval",
default=False,
action='store_true',
help="Whether to run eval on the dev set.")
parser.add_argument("--train_batch_size",
default=32,
type=int,
help="Total batch size for training.")
parser.add_argument("--eval_batch_size",
default=8,
type=int,
help="Total batch size for eval.")
parser.add_argument("--learning_rate",
default=5e-5,
type=float,
help="The initial learning rate for Adam.")
parser.add_argument("--num_train_epochs",
default=3.0,
type=float,
help="Total number of training epochs to perform.")
parser.add_argument("--warmup_proportion",
default=0.1,
type=float,
help="Proportion of training to perform linear learning rate warmup for. "
"E.g., 0.1 = 10%% of training.")
parser.add_argument("--no_cuda",
default=False,
action='store_true',
help="Whether not to use CUDA when available")
parser.add_argument("--local_rank",
type=int,
default=-1,
help="local_rank for distributed training on gpus")
parser.add_argument('--seed',
type=int,
default=42,
help="random seed for initialization")
parser.add_argument('--gradient_accumulation_steps',
type=int,
default=1,
help="Number of updates steps to accumualte before performing a backward/update pass.")
experiment_name = 'horror_uncased_5_tied_mlm'
argv = """
--task_name lm \
--data_dir {DATA_DIR} \
--vocab_file {BERT_BASE_DIR}/vocab.txt \
--bert_config_file {BERT_BASE_DIR}/bert_config.json \
--init_checkpoint {BERT_BASE_DIR}/pytorch_model.bin \
--do_train \
--do_eval \
--gradient_accumulation_steps 2 \
--train_batch_size 32 \
--learning_rate 3e-5 \
--num_train_epochs 3.0 \
--max_seq_length 128 \
--output_dir ../outputs/{name}/
""".format(
BERT_BASE_DIR='../data/weights/cased_L-12_H-768_A-12',
DATA_DIR='../data/input/horror_gutenberg',
name=experiment_name
).replace('\n', '').split(' ')
print(argv)
args = parser.parse_args(argv)
if args.local_rank == -1 or args.no_cuda:
device = torch.device("cuda" if torch.cuda.is_available() and not args.no_cuda else "cpu")
n_gpu = torch.cuda.device_count()
else:
device = torch.device("cuda", args.local_rank)
n_gpu = 1
# Initializes the distributed backend which will take care of sychronizing nodes/GPUs
torch.distributed.init_process_group(backend='nccl')
logger.info("device %s n_gpu %d distributed training %r", device, n_gpu, bool(args.local_rank != -1))
if args.gradient_accumulation_steps < 1:
raise ValueError("Invalid gradient_accumulation_steps parameter: {}, should be >= 1".format(
args.gradient_accumulation_steps))
args.train_batch_size = int(args.train_batch_size / args.gradient_accumulation_steps)
random.seed(args.seed)
np.random.seed(args.seed)
torch.manual_seed(args.seed)
if n_gpu > 0:
torch.cuda.manual_seed_all(args.seed)
if not args.do_train and not args.do_eval:
raise ValueError("At least one of `do_train` or `do_eval` must be True.")
bert_config = BertConfig.from_json_file(args.bert_config_file)
if args.max_seq_length > bert_config.max_position_embeddings:
raise ValueError(
"Cannot use sequence length {} because the BERT model was only trained up to sequence length {}".format(
args.max_seq_length, bert_config.max_position_embeddings))
if os.path.exists(args.output_dir) and os.listdir(args.output_dir):
print("Output directory ({}) already exists and is not empty.".format(args.output_dir))
os.makedirs(args.output_dir, exist_ok=True)
save_path = os.path.join(args.output_dir, 'state_dict.pkl')
save_path
tokenizer = tokenization.FullTokenizer(
vocab_file=args.vocab_file, do_lower_case=args.do_lower_case)
decoder = {v:k for k,v in tokenizer.wordpiece_tokenizer.vocab.items()}
processors = {
"lm": LMProcessor,
}
task_name = args.task_name.lower()
if task_name not in processors:
raise ValueError("Task not found: %s" % (task_name))
processor = processors[task_name](tokenizer=tokenizer)
label_list = processor.get_labels()
train_examples = processor.get_train_examples(args.data_dir, skip=30, tqdm=tqdm)
num_train_steps = int(
len(train_examples) / args.train_batch_size * args.num_train_epochs)
train_features = convert_tokens_to_features(
train_examples, label_list, args.max_seq_length, tokenizer, tqdm=tqdm)
all_input_ids = torch.tensor([f.input_ids for f in train_features], dtype=torch.long)
all_input_mask = torch.tensor([f.input_mask for f in train_features], dtype=torch.long)
all_segment_ids = torch.tensor([f.segment_ids for f in train_features], dtype=torch.long)
all_label_ids = torch.tensor([f.label_id for f in train_features], dtype=torch.long)
all_label_weights = torch.tensor([f.label_weights for f in train_features], dtype=torch.long)
train_data = TensorDataset(all_input_ids, all_input_mask, all_segment_ids, all_label_ids, all_label_weights)
if args.local_rank == -1:
train_sampler = RandomSampler(train_data)
else:
train_sampler = DistributedSampler(train_data)
train_dataloader = DataLoader(train_data, sampler=train_sampler, batch_size=args.train_batch_size)
model = BertForMaskedLanguageModelling(bert_config)
if args.init_checkpoint is not None:
model.bert.load_state_dict(torch.load(args.init_checkpoint, map_location='cpu'))
if os.path.isfile(save_path):
model.load_state_dict(torch.load(save_path, map_location='cpu'))
model.to(device)
if args.local_rank != -1:
model = torch.nn.parallel.DistributedDataParallel(model, device_ids=[args.local_rank],
output_device=args.local_rank)
elif n_gpu > 1:
model = torch.nn.DataParallel(model)
model
no_decay = ['bias', 'gamma', 'beta']
optimizer_parameters = [
{'params': [p for n, p in model.named_parameters() if n not in no_decay], 'weight_decay_rate': 0.01},
{'params': [p for n, p in model.named_parameters() if n in no_decay], 'weight_decay_rate': 0.0}
]
optimizer = BERTAdam(optimizer_parameters,
lr=args.learning_rate,
warmup=args.warmup_proportion,
t_total=num_train_steps)
val_test="""Another gentleman has fallen a victim to the terrible epidemic of suicide which for the last month has prevailed in the West End. Mr. Sidney Crashaw, of Stoke House, Fulham, and King's Pomeroy, Devon, was found, after a prolonged search, hanging dead from the branch of a tree in his garden at one o'clock today. The deceased gentleman dined last night at the Carlton Club and seemed in his usual health and spirits. He left the club at about ten o'clock, and was seen walking leisurely up St. James's Street a little later. Subsequent to this his movements cannot be traced"""
display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=30, T=1, device=device))
display(predict_masked_words(val_test, processor, tokenizer, model, device=device, max_seq_length=args.max_seq_length))
global_step = 0
1/0
model.train()
for _ in tqdm(range(int(args.num_train_epochs)), desc="Epoch"):
tr_loss, nb_tr_examples, nb_tr_steps = 0, 0, 0
with tqdm(total=len(train_dataloader), desc='Iteration', mininterval=0.5) as prog:
for step, batch in enumerate(train_dataloader):
batch = tuple(t.to(device) for t in batch)
input_ids, input_mask, segment_ids, label_ids, label_weights = batch
loss, logits = model(input_ids, segment_ids, input_mask, label_ids, label_weights)
if n_gpu > 1:
loss = loss.mean() # mean() to average on multi-gpu.
if args.gradient_accumulation_steps > 1:
loss = loss / args.gradient_accumulation_steps
loss.backward()
tr_loss += loss.item()
nb_tr_examples += input_ids.size(0)
nb_tr_steps += 1
if (step + 1) % args.gradient_accumulation_steps == 0:
optimizer.step() # We have accumulated enougth gradients
model.zero_grad()
prog.update(1)
prog.desc = 'Iter. loss={:2.6f}'.format(tr_loss/nb_tr_examples)
if step%3000==10:
print('step', step, 'loss', tr_loss/nb_tr_examples)
display(predict_masked_words(val_test, processor, tokenizer, model, device=device, max_seq_length=args.max_seq_length))
display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=10, device=device))
tr_loss, nb_tr_examples, nb_tr_steps = 0, 0, 0
torch.save(model.state_dict(), save_path)
global_step += 1
torch.save(model.state_dict(), save_path)
# val_test="""Another gentleman has fallen a victim to the terrible epidemic of suicide which for the last month has prevailed in the West End. Mr. Sidney Crashaw, of Stoke House, Fulham, and King's Pomeroy, Devon, was found, after a prolonged search, hanging dead from the branch of a tree in his garden at one o'clock today. The deceased gentleman dined last night at the Carlton Club and seemed in his usual health and spirits. He left the club at about ten o'clock, and was seen walking leisurely up St. James's Street a little later. Subsequent to this his movements cannot be traced. On the discovery of the body medical aid was at once summoned, but life had evidently been long extinct. So far as is known, Mr. Crashaw had no trouble or anxiety of any kind. This painful suicide, it will be remembered, is the fifth of the kind in the last month. The authorities at Scotland Yard are unable to suggest any explanation of these terrible occurrences."""
# display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=100, T=.5, device=device))
val_test="""His mind spun in on itself . . . ."""
display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=150, T=.2, device=device))
val_test="""A giant spider descended on to . . . ."""
display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=100, T=.5, device=device))
val_test="""Madness enveloped his mind as . . . ."""
display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=100, T=.5, device=device))
val_test="""A thin film of . . . ."""
display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=100, T=.01, device=device))
val_test="""Quivering with fear, he trembled as . . . ."""
display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=100, T=.7, device=device))
val_test="""Madness enveloped his mind as . . . ."""
display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=128, T=.1, device=device))
val_test="""All at once, in a moment of realisation, he knew the secret to creating true artificial intelligence was . . . ."""
display(predict_next_words(val_test, processor, tokenizer, model, max_seq_length=args.max_seq_length, n=300, T=.5, device=device))
improve_words_recursive(text, processor, tokenizer, model, iterations=100, max_seq_length=128, n=10, T=0.3, device="cuda", debug=10)
| 0.620852 | 0.489626 |
```
#!/usr/bin/env python
# coding: utf-8
# In[31]:
import re
import matplotlib.pyplot as plt
import pylab as pl
import numpy as np
import math
import os
from collections import defaultdict
from matplotlib.patches import Rectangle
from matplotlib.patches import Rectangle
import numpy as np
import pylab as P
import random
# In[242]:
def compute_exp(path):
regex = re.compile(r"\d+:(\d+):\s+(\d+\.\d+):(\d+\.\d+)")
obs_lst = []
ref_lst = []
with open(os.path.join(path, 'monte_carlo_sampling_obs.txt')) as f:
for line in f:
line = line.rstrip('\n')
#print(line)
r = re.search(regex, line)
#print(r)
if r is not None:
obs_lst.append(float(r.group(3)))
else:
print(line)
with open(os.path.join(path, 'monte_carlo_sampling_ref.txt')) as f:
for line in f:
line = line.rstrip('\n')
r = re.search(regex, line)
if r is not None:
ref_lst.append(float(r.group(3)))
else:
print(line)
#print(random_count_lst_dct)
return np.asarray(obs_lst), np.asarray(ref_lst)
# In[176]:
def plot_box_plt(yobs, yref, fig_name):
d_lst = list(pl.frange(0.1,0.5,0.01))
eps = 0.05 #controls amount of jitter
xobs = [random.uniform(1-eps,1+eps) for i in range(0,yobs.shape[0])]
xref = [random.uniform(2-eps,2+eps) for i in range(0,yref.shape[0])]
box_data = [yobs, yref]
plt.plot(xobs, yobs, 'ro', label=r'$\mathcal{R}_{obs}$')
plt.plot(xref, yref, 'bo', label=r'$\mathcal{R}_{ref}$')
xnames = [r'$\mathcal{R}_{obs}$', r'$\mathcal{R}_{ref}$']
plt.boxplot(box_data,labels=xnames,sym="") #dont show outliers
from matplotlib import rcParams
labelsize = 24
rcParams['xtick.labelsize'] = labelsize
#plt.yticks([])
plt.yticks(np.arange(0.1, 0.4, 0.05))
plt.yticks([])
plt.legend(loc='center left', shadow=True, facecolor='white', framealpha=1, prop={'size': 16})
plt.savefig(fig_name, bbox_inches='tight', dpi =800, pad_inches=0)
plt.show() # render pipeline
plt.close()
# In[150]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_10m_celebahq/monte_carlo_sampling')
print(yobs)
plot_box_plt(yobs, yref, 'boxplot_SGAN_CelebAHQ_1024.pdf')
# In[177]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_10m_ffhq/monte_carlo_sampling')
print(yobs)
yref = list(yref)
yref.remove(max(yref))
yref.remove(max(yref))
plot_box_plt(yobs, np.asarray(yref), 'boxplot_SGAN_FFHQ_1024.pdf')
# In[152]:
yobs, yref = compute_exp('legacy/pggan/monte_carlo_sampling_10m_ffhq/monte_carlo_sampling')
print(yobs)
plot_box_plt(yobs, yref, 'boxplot_PGGAN_FFHQ_1024.pdf')
# In[154]:
yobs, yref = compute_exp('legacy/pggan/monte_carlo_sampling_10m_celebahq/monte_carlo_sampling')
print(yobs)
plot_box_plt(yobs, yref, 'boxplot_PGGAN_CelebAHQ_1024.pdf')
# In[ ]:
import random
# In[169]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_1m_finetune/monte_carlo_sampling')
plot_box_plt(yobs[random.sample(range(50), 8)], yref, 'boxplot_SGAN_Finetune_128.pdf')
# In[173]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_1m_randomness/monte_carlo_sampling')
plot_box_plt(yobs[random.sample(range(100), 5)], yref, 'boxplot_SGAN_Randomness_128.pdf')
# In[159]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_1m_sgan_architecture/monte_carlo_sampling')
print(yobs)
plot_box_plt(yobs[random.sample(range(100), 4)], yref, 'boxplot_SGAN_Architecture_128.pdf')
# In[ ]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_1m_pggan_architecture/monte_carlo_sampling')
print(yobs)
yref = list(yref)
yref.remove(max(yref))
plot_box_plt(yobs[random.sample(range(100), 4)], np.asarray(yref), 'boxplot_PGGAN_Architecture_128.pdf')
# In[236]:
def plot_2box_plt(yobs1, yref1, yobs2, yref2, fig_name):
d_lst = list(pl.frange(0.1,0.5,0.01))
eps = 0.05 #controls amount of jitter
xobs1 = [random.uniform(-0.4-eps,-0.4+eps) for i in range(0,yobs1.shape[0])]
xref1 = [random.uniform(0.4-eps,0.4+eps) for i in range(0,yref1.shape[0])]
box_data1 = [yobs1, yobs2]
xobs2 = [random.uniform(1.6-eps,1.6+eps) for i in range(0,yobs2.shape[0])]
xref2 = [random.uniform(2.4-eps,2.4+eps) for i in range(0,yref2.shape[0])]
box_data2 = [yref1, yref2]
plt.plot(xobs1, yobs1, 'ro', label=r'$\mathcal{R}_{obs}$')
plt.plot(xref1, yref1, 'bo', label=r'$\mathcal{R}_{ref}$')
plt.plot(xobs2, yobs2, 'ro')
plt.plot(xref2, yref2, 'bo')
xnames = ['Before Calibration', 'After Calibration']
#plt.boxplot(box_data1,positions = [-0.4, 1.6],labels=xnames,sym="", widths=0.6)
#plt.boxplot(box_data2,positions = [0.4, 2.4],labels=xnames,sym="", widths=0.6)
plt.boxplot(box_data1,positions = [-0.4, 1.6],sym="", widths=0.6)
plt.boxplot(box_data2,positions = [0.4, 2.4],sym="", widths=0.6)
plt.xticks(range(0, len(xnames) * 2, 2), xnames)
plt.xlim(-2, len(xnames)*2)
#plt.ylim(0, 8)
plt.tight_layout()
from matplotlib import rcParams
labelsize = 12
rcParams['xtick.labelsize'] = labelsize
plt.yticks(np.arange(0.1, 0.4, 0.05))
#plt.yticks([])
plt.legend(loc='center left', shadow=True, facecolor='white', framealpha=1, prop={'size': 16})
plt.savefig(fig_name, bbox_inches='tight', dpi =800, pad_inches=0)
plt.show() # render pipeline
plt.close()
# In[260]:
yobs1, yref1 = compute_exp('legacy/pggan/monte_carlo_sampling_10m_celebahq/monte_carlo_sampling')
yobs2, yref2 = compute_exp('legacy/pggan/monte_carlo_sampling_10m_ffhq/monte_carlo_sampling')
print(yobs)
plot_2box_plt(np.asarray([np.min(yobs2)]), yref2, np.asarray([np.max(yobs1)]), yref1, 'boxplot_GMM_reshaping.pdf')
# In[261]:
yobs1, yref1 = compute_exp('legacy/pggan/monte_carlo_sampling_10m_ffhq/monte_carlo_sampling')
yobs2, yref2 = compute_exp('legacy/pggan/monte_carlo_sampling_10m_ffhq/monte_carlo_sampling_old')
print(yobs)
plot_2box_plt(np.asarray([np.min(yobs2)]), yref2, np.asarray([np.mean(yobs1)]), yref1, 'boxplot_IS_reshaping.pdf')
```
|
github_jupyter
|
#!/usr/bin/env python
# coding: utf-8
# In[31]:
import re
import matplotlib.pyplot as plt
import pylab as pl
import numpy as np
import math
import os
from collections import defaultdict
from matplotlib.patches import Rectangle
from matplotlib.patches import Rectangle
import numpy as np
import pylab as P
import random
# In[242]:
def compute_exp(path):
regex = re.compile(r"\d+:(\d+):\s+(\d+\.\d+):(\d+\.\d+)")
obs_lst = []
ref_lst = []
with open(os.path.join(path, 'monte_carlo_sampling_obs.txt')) as f:
for line in f:
line = line.rstrip('\n')
#print(line)
r = re.search(regex, line)
#print(r)
if r is not None:
obs_lst.append(float(r.group(3)))
else:
print(line)
with open(os.path.join(path, 'monte_carlo_sampling_ref.txt')) as f:
for line in f:
line = line.rstrip('\n')
r = re.search(regex, line)
if r is not None:
ref_lst.append(float(r.group(3)))
else:
print(line)
#print(random_count_lst_dct)
return np.asarray(obs_lst), np.asarray(ref_lst)
# In[176]:
def plot_box_plt(yobs, yref, fig_name):
d_lst = list(pl.frange(0.1,0.5,0.01))
eps = 0.05 #controls amount of jitter
xobs = [random.uniform(1-eps,1+eps) for i in range(0,yobs.shape[0])]
xref = [random.uniform(2-eps,2+eps) for i in range(0,yref.shape[0])]
box_data = [yobs, yref]
plt.plot(xobs, yobs, 'ro', label=r'$\mathcal{R}_{obs}$')
plt.plot(xref, yref, 'bo', label=r'$\mathcal{R}_{ref}$')
xnames = [r'$\mathcal{R}_{obs}$', r'$\mathcal{R}_{ref}$']
plt.boxplot(box_data,labels=xnames,sym="") #dont show outliers
from matplotlib import rcParams
labelsize = 24
rcParams['xtick.labelsize'] = labelsize
#plt.yticks([])
plt.yticks(np.arange(0.1, 0.4, 0.05))
plt.yticks([])
plt.legend(loc='center left', shadow=True, facecolor='white', framealpha=1, prop={'size': 16})
plt.savefig(fig_name, bbox_inches='tight', dpi =800, pad_inches=0)
plt.show() # render pipeline
plt.close()
# In[150]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_10m_celebahq/monte_carlo_sampling')
print(yobs)
plot_box_plt(yobs, yref, 'boxplot_SGAN_CelebAHQ_1024.pdf')
# In[177]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_10m_ffhq/monte_carlo_sampling')
print(yobs)
yref = list(yref)
yref.remove(max(yref))
yref.remove(max(yref))
plot_box_plt(yobs, np.asarray(yref), 'boxplot_SGAN_FFHQ_1024.pdf')
# In[152]:
yobs, yref = compute_exp('legacy/pggan/monte_carlo_sampling_10m_ffhq/monte_carlo_sampling')
print(yobs)
plot_box_plt(yobs, yref, 'boxplot_PGGAN_FFHQ_1024.pdf')
# In[154]:
yobs, yref = compute_exp('legacy/pggan/monte_carlo_sampling_10m_celebahq/monte_carlo_sampling')
print(yobs)
plot_box_plt(yobs, yref, 'boxplot_PGGAN_CelebAHQ_1024.pdf')
# In[ ]:
import random
# In[169]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_1m_finetune/monte_carlo_sampling')
plot_box_plt(yobs[random.sample(range(50), 8)], yref, 'boxplot_SGAN_Finetune_128.pdf')
# In[173]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_1m_randomness/monte_carlo_sampling')
plot_box_plt(yobs[random.sample(range(100), 5)], yref, 'boxplot_SGAN_Randomness_128.pdf')
# In[159]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_1m_sgan_architecture/monte_carlo_sampling')
print(yobs)
plot_box_plt(yobs[random.sample(range(100), 4)], yref, 'boxplot_SGAN_Architecture_128.pdf')
# In[ ]:
yobs, yref = compute_exp('legacy/sgan/monte_carlo_sampling_1m_pggan_architecture/monte_carlo_sampling')
print(yobs)
yref = list(yref)
yref.remove(max(yref))
plot_box_plt(yobs[random.sample(range(100), 4)], np.asarray(yref), 'boxplot_PGGAN_Architecture_128.pdf')
# In[236]:
def plot_2box_plt(yobs1, yref1, yobs2, yref2, fig_name):
d_lst = list(pl.frange(0.1,0.5,0.01))
eps = 0.05 #controls amount of jitter
xobs1 = [random.uniform(-0.4-eps,-0.4+eps) for i in range(0,yobs1.shape[0])]
xref1 = [random.uniform(0.4-eps,0.4+eps) for i in range(0,yref1.shape[0])]
box_data1 = [yobs1, yobs2]
xobs2 = [random.uniform(1.6-eps,1.6+eps) for i in range(0,yobs2.shape[0])]
xref2 = [random.uniform(2.4-eps,2.4+eps) for i in range(0,yref2.shape[0])]
box_data2 = [yref1, yref2]
plt.plot(xobs1, yobs1, 'ro', label=r'$\mathcal{R}_{obs}$')
plt.plot(xref1, yref1, 'bo', label=r'$\mathcal{R}_{ref}$')
plt.plot(xobs2, yobs2, 'ro')
plt.plot(xref2, yref2, 'bo')
xnames = ['Before Calibration', 'After Calibration']
#plt.boxplot(box_data1,positions = [-0.4, 1.6],labels=xnames,sym="", widths=0.6)
#plt.boxplot(box_data2,positions = [0.4, 2.4],labels=xnames,sym="", widths=0.6)
plt.boxplot(box_data1,positions = [-0.4, 1.6],sym="", widths=0.6)
plt.boxplot(box_data2,positions = [0.4, 2.4],sym="", widths=0.6)
plt.xticks(range(0, len(xnames) * 2, 2), xnames)
plt.xlim(-2, len(xnames)*2)
#plt.ylim(0, 8)
plt.tight_layout()
from matplotlib import rcParams
labelsize = 12
rcParams['xtick.labelsize'] = labelsize
plt.yticks(np.arange(0.1, 0.4, 0.05))
#plt.yticks([])
plt.legend(loc='center left', shadow=True, facecolor='white', framealpha=1, prop={'size': 16})
plt.savefig(fig_name, bbox_inches='tight', dpi =800, pad_inches=0)
plt.show() # render pipeline
plt.close()
# In[260]:
yobs1, yref1 = compute_exp('legacy/pggan/monte_carlo_sampling_10m_celebahq/monte_carlo_sampling')
yobs2, yref2 = compute_exp('legacy/pggan/monte_carlo_sampling_10m_ffhq/monte_carlo_sampling')
print(yobs)
plot_2box_plt(np.asarray([np.min(yobs2)]), yref2, np.asarray([np.max(yobs1)]), yref1, 'boxplot_GMM_reshaping.pdf')
# In[261]:
yobs1, yref1 = compute_exp('legacy/pggan/monte_carlo_sampling_10m_ffhq/monte_carlo_sampling')
yobs2, yref2 = compute_exp('legacy/pggan/monte_carlo_sampling_10m_ffhq/monte_carlo_sampling_old')
print(yobs)
plot_2box_plt(np.asarray([np.min(yobs2)]), yref2, np.asarray([np.mean(yobs1)]), yref1, 'boxplot_IS_reshaping.pdf')
| 0.250638 | 0.464659 |
# Gráficos com Matplotlib
```
%matplotlib inline
import pandas as pd
import matplotlib.pyplot as plt
plt.rc('figure', figsize=(15,8))
dados = pd.read_csv('../dados/aluguel2.csv', sep=';')
dados.head()
```
## Criar uma área com vários gráficos
```
area = plt.figure()
```
### Colocando 4 gráficos dentro dessa área
```
# sigfinica que na área terão 4 gráficos
# 2 linhas, 2 colunas e este estará na posição 1
g1 = area.add_subplot(2, 2, 1)
# fazendo para os outros gráficos
g2 = area.add_subplot(2, 2, 2)
g3 = area.add_subplot(2, 2, 3)
g4 = area.add_subplot(2, 2, 4)
```
### Criando gráficos de dispersão
```
# cria um gráfico de dispersão, com as variáveis passadas
g1.scatter(dados.Valor, dados.Area)
# cria um título para o gráfico
g1.set_title('Valor x Área')
area
```
### Criando um gráfico de histograma
```
g2.hist(dados.Valor)
g2.set_title('Histograma')
```
### Criando um amostra aleatória dentro do próprio dataframe
```
# cria-se uma amostra aleatória com 100 registros
dados_g3 = dados.Valor.sample(100)
# reconstruindo o index
dados_g3.index = range(dados_g3.shape[0])
# passa esses dados para o gráfico
g3.plot(dados_g3)
# lembrando que cada vez que a célula for rodada, será criado uma amostra, portando um gráfico diferente
g3.set_title('Amostra (Valor)')
```
### Criando um gráfico de barras
```
# agrupando por tipo e pegando apenas os valores
grupo = dados.groupby('Tipo')['Valor']
# criando os rótulos
label = grupo.mean().index
valores = grupo.mean().values
# o gráfico de barras precisa dos rótulos e valores
g4.bar(label, valores)
g4.set_title('Valor Médio por Tipo')
```
### Visualizando todos os gráficos
* para deixar essas áreas vazias é possível usar:
`area = ''`
```
# visualiza todos os gráficos gerados na área previamente configurada
area
```
## Salvando a imagem gerada
```
# define um nome para a figura
# ajusta a definição da imagem
# estreita as margens brancas entre gráficos para melhor colagem da imagem
area.savefig('../dados/grafico.png', dpi=300, bbox_inches='tight')
```
### Rodando a imagem em Markdown
``

## Exercício
### Considere neste exercício o arquivo aluguel_amostra.csv e indique qual o código necessário para gerar os gráficos da figura a seguir:

* Neste exercício, estamos apresentando o gráfico de pizza que pode ser obtido com a aplicação do método pie(), de matplotlib.
* Considere o código inicial abaixo para resolver o exercício:
```
%matplotlib inline
import pandas as pd
import matplotlib.pyplot as plt
plt.rc('figure', figsize = (15, 7))
dados = pd.read_csv('dados/aluguel_amostra.csv', sep = ';')
```
```
%matplotlib inline
import pandas as pd
import matplotlib.pyplot as plt
plt.rc('figure', figsize = (15, 7))
dados_exercicio = pd.read_csv('../dados/aluguel_amostra.csv', sep = ';')
dados_exercicio.head()
# cria a área
area = plt.figure()
# cria e dimensiona a variável dos gráficos
g1 = area.add_subplot(1, 2, 1)
g2 = area.add_subplot(1, 2, 2)
# seleciona os valores e rótulos
grupo1 = dados.groupby('Tipo Agregado')['Valor']
label = grupo1.count().index
valores = grupo1.count().values
# passa os parâmetros para o gráfico e ajusta o valor exibido para % com uma casa decimal
g1.pie(valores, labels = label, autopct='%.1f%%')
# define o título
g1.set_title('Total de Imóveis por Tipo Agregado')
# seleciona os valores e rótulos
grupo2 = dados.groupby('Tipo')['Valor']
label = grupo2.count().index
valores = grupo2.count().values
# passa os parâmetros para o gráfico e ajusta o valor exibido para % com uma casa decimal
# explode descola os valores do gráfico
g2.pie(valores, labels = label, autopct='%.1f%%', explode = (.1, .1, .1, .1, .1))
# define o título
g2.set_title('Total de Imóveis por Tipo')
```
## Extra com gráfico de pizza
### Analisando a variável bairros
```
dados_exercicio['Bairro'].unique()
```
### Selecionando somente alguns bairros
```
bairros = ['Copacabana', 'Leblon', 'Flamengo', 'Botafogo', 'Lapa']
selecao = dados_exercicio['Bairro'].isin(bairros)
dados_exercicio = dados_exercicio[selecao]
dados_exercicio.head()
# note que agora o dataframe contem apenas os bairros da variável bairros
```
### Agrupando os bairros
```
dados_exercicio.Bairro.value_counts()
```
### Definindo valores e rótulos do grafico de pizza
* gráficos de pizza recebem como valores apenas um array 1D
* portanto, selecionaremos apenas os valores da Series acima
```
valores = dados_exercicio.Bairro.value_counts().values
valores
```
* como rótulos passaremos o index da Series
```
label = dados_exercicio.Bairro.value_counts().index
label
```
### Plotando o gráfico
#### 1º Método
```
plt.pie(valores, labels=label, autopct='%.2f%%', explode=(.1, .1, .1, .1, .1))
```
#### 2º Método
```
# define a área onde será plotado
grafico = plt.figure()
# cria um objeto do tipo sub-grafico
grafico_pizza = grafico.add_subplot()
# aplica para esse objeto o grafico de pizza
# com %, 2 casas decimais, e descola os valores
grafico_pizza.pie(valores, labels=label, autopct='%.2f%%', explode=(.1, .1, .1, .1, .1))
# acerta o título e o tamanho do título em extra grande
grafico_pizza.set_title('Bairros Mais Conhecidos', size='xx-large')
```
|
github_jupyter
|
%matplotlib inline
import pandas as pd
import matplotlib.pyplot as plt
plt.rc('figure', figsize=(15,8))
dados = pd.read_csv('../dados/aluguel2.csv', sep=';')
dados.head()
area = plt.figure()
# sigfinica que na área terão 4 gráficos
# 2 linhas, 2 colunas e este estará na posição 1
g1 = area.add_subplot(2, 2, 1)
# fazendo para os outros gráficos
g2 = area.add_subplot(2, 2, 2)
g3 = area.add_subplot(2, 2, 3)
g4 = area.add_subplot(2, 2, 4)
# cria um gráfico de dispersão, com as variáveis passadas
g1.scatter(dados.Valor, dados.Area)
# cria um título para o gráfico
g1.set_title('Valor x Área')
area
g2.hist(dados.Valor)
g2.set_title('Histograma')
# cria-se uma amostra aleatória com 100 registros
dados_g3 = dados.Valor.sample(100)
# reconstruindo o index
dados_g3.index = range(dados_g3.shape[0])
# passa esses dados para o gráfico
g3.plot(dados_g3)
# lembrando que cada vez que a célula for rodada, será criado uma amostra, portando um gráfico diferente
g3.set_title('Amostra (Valor)')
# agrupando por tipo e pegando apenas os valores
grupo = dados.groupby('Tipo')['Valor']
# criando os rótulos
label = grupo.mean().index
valores = grupo.mean().values
# o gráfico de barras precisa dos rótulos e valores
g4.bar(label, valores)
g4.set_title('Valor Médio por Tipo')
# visualiza todos os gráficos gerados na área previamente configurada
area
# define um nome para a figura
# ajusta a definição da imagem
# estreita as margens brancas entre gráficos para melhor colagem da imagem
area.savefig('../dados/grafico.png', dpi=300, bbox_inches='tight')
%matplotlib inline
import pandas as pd
import matplotlib.pyplot as plt
plt.rc('figure', figsize = (15, 7))
dados = pd.read_csv('dados/aluguel_amostra.csv', sep = ';')
%matplotlib inline
import pandas as pd
import matplotlib.pyplot as plt
plt.rc('figure', figsize = (15, 7))
dados_exercicio = pd.read_csv('../dados/aluguel_amostra.csv', sep = ';')
dados_exercicio.head()
# cria a área
area = plt.figure()
# cria e dimensiona a variável dos gráficos
g1 = area.add_subplot(1, 2, 1)
g2 = area.add_subplot(1, 2, 2)
# seleciona os valores e rótulos
grupo1 = dados.groupby('Tipo Agregado')['Valor']
label = grupo1.count().index
valores = grupo1.count().values
# passa os parâmetros para o gráfico e ajusta o valor exibido para % com uma casa decimal
g1.pie(valores, labels = label, autopct='%.1f%%')
# define o título
g1.set_title('Total de Imóveis por Tipo Agregado')
# seleciona os valores e rótulos
grupo2 = dados.groupby('Tipo')['Valor']
label = grupo2.count().index
valores = grupo2.count().values
# passa os parâmetros para o gráfico e ajusta o valor exibido para % com uma casa decimal
# explode descola os valores do gráfico
g2.pie(valores, labels = label, autopct='%.1f%%', explode = (.1, .1, .1, .1, .1))
# define o título
g2.set_title('Total de Imóveis por Tipo')
dados_exercicio['Bairro'].unique()
bairros = ['Copacabana', 'Leblon', 'Flamengo', 'Botafogo', 'Lapa']
selecao = dados_exercicio['Bairro'].isin(bairros)
dados_exercicio = dados_exercicio[selecao]
dados_exercicio.head()
# note que agora o dataframe contem apenas os bairros da variável bairros
dados_exercicio.Bairro.value_counts()
valores = dados_exercicio.Bairro.value_counts().values
valores
label = dados_exercicio.Bairro.value_counts().index
label
plt.pie(valores, labels=label, autopct='%.2f%%', explode=(.1, .1, .1, .1, .1))
# define a área onde será plotado
grafico = plt.figure()
# cria um objeto do tipo sub-grafico
grafico_pizza = grafico.add_subplot()
# aplica para esse objeto o grafico de pizza
# com %, 2 casas decimais, e descola os valores
grafico_pizza.pie(valores, labels=label, autopct='%.2f%%', explode=(.1, .1, .1, .1, .1))
# acerta o título e o tamanho do título em extra grande
grafico_pizza.set_title('Bairros Mais Conhecidos', size='xx-large')
| 0.249539 | 0.944074 |
# Polynomial Regression
The correlation between the variables are not always linear, to handle these cases we use polynomials to represent the realtions
### Aim
To find the price of a speaker system based on the required output power.
### Data
The data used in this example is generated in using a python program.
The Data is about the **price in Euros** of a speaker and the sound **output in Watts** of the same.
**Note** : The data used generated using a python program.
### Libraries Used
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy import stats
from sklearn.metrics import r2_score
from pylab import *
%matplotlib inline
```
### Loading the Data
```
# Data saved in the same folder
df = pd.read_csv("Data/SpeakerPrice.csv")
df.columns
```
The Dataset contains Name, Price and Power in watts, We can ignore the values in the column **Name** in this use-case
### Plot of 'Power Vs Price'
We are checking if any correlation exists between power and the price of the speakers
```
scatter(df['Price'] , df['Watt'] )
# Adding labels
plt.xlabel("Price (in Euros)")
plt.ylabel("Power (in Watts)")
```
#### Observation :
we can observe that there is a non-linear correlation between Price and Power. <br>
In the data set, we can observe as the power rating **increases**, the variance in the inflation of the price **decreases**. <br> Hence a polynomial regression has to be used to predict the price based on the power.
#### Polynomial function :
Creating a polynomial funtion with **degree 2** to **degree 7** to find the which degree polynomial is optimal
```
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 2))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 2 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 3))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 3 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 4))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 4 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 5))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 5 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 6))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 6 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 7))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 7 : " , r2)
```
#### **Observation** :
The improvement we achieve by increasing the **degree** of the polynomial from **4** and above do not yeild much improvement in **r2 score**. <br>
Hence the **Polynomial degree = 5** will be set for this dataset
### Plotting
Fitting a polynomial function and plotting the same
```
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 4))
### plotting the polynomial Function (red line)
ls = np.linspace(0, 10000, 1000)
### the values of Price vs Power is added to the grapgh
plt.scatter(df['Price'], df['Watt'])
### Labels
plt.xlabel("Price (in Euros)")
plt.ylabel("Power (in Watts)")
### the Line is drawn
plt.plot(ls, Line(ls), c='r')
### printing the graph
plt.show()
```
### Polynomial Function used
```
Line
```
This represents the following polynomial function <br>
**-4.60905628e-13 * x ^ 4 + 1.10984448e-08 * x ^ 3 + -9.61617248e-05 * x ^ 2 + 4.03585669e-01 * x + 6.10130389e+01**
### Prediction
A customer want to check the estimated price of a system with a power requirement of **5000 Watts**
```
power=5000
estimated_price = Line(power)
print(estimated_price)
```
### Conclusion
According to the Current model,
the approximate price to be expected is **~769.80 Euro** <br>
for a new speaker with **power 5000 Watts**
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy import stats
from sklearn.metrics import r2_score
from pylab import *
%matplotlib inline
# Data saved in the same folder
df = pd.read_csv("Data/SpeakerPrice.csv")
df.columns
scatter(df['Price'] , df['Watt'] )
# Adding labels
plt.xlabel("Price (in Euros)")
plt.ylabel("Power (in Watts)")
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 2))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 2 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 3))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 3 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 4))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 4 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 5))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 5 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 6))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 6 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 7))
r2 = r2_score(df['Watt'], Line(df['Price']))
print("R2 vlaue for Polynomial Function with degree 7 : " , r2)
Line = np.poly1d(np.polyfit(df['Price'], df['Watt'], 4))
### plotting the polynomial Function (red line)
ls = np.linspace(0, 10000, 1000)
### the values of Price vs Power is added to the grapgh
plt.scatter(df['Price'], df['Watt'])
### Labels
plt.xlabel("Price (in Euros)")
plt.ylabel("Power (in Watts)")
### the Line is drawn
plt.plot(ls, Line(ls), c='r')
### printing the graph
plt.show()
Line
power=5000
estimated_price = Line(power)
print(estimated_price)
| 0.565779 | 0.99269 |
# Lecture 15: Other file formats
CSCI 1360: Foundations for Informatics and Analytics
## Overview and Objectives
In the last lecture, we looked at some ways of interacting with the filesystem through Python and how to read data off files stored on the hard drive. We looked at raw text files; however, there are numerous structured formats that these files can take, and we'll explore some of those here. By the end of this lecture, you should be able to:
- Identify some of the primary data storage formats
- Explain how to use other tools for some of the more exotic data types
## Part 1: Comma-separated value (CSV) files
We've discussed text formats: each line of text in a file can be treated as a string in a list of strings. What else might we encounter in our data science travels?
Easily the most common text file format is the CSV, or comma-separated values format. This is pretty much what it sounds like: if you have (semi-) structured data, you can delineate spaces between data using commas (or, to generalize, other characters like tabs).
As an example, we could represent a matrix very easily using the CSV format. The file storing a 3x3 matrix would look something like this:
<pre>
1,2,3
4,5,6
7,8,9
</pre>
Each row is on one line by itself, and the columns are separated by commas.
How can we read a CSV file? One way, potentially, is just do it yourself:
```
# File "csv_file.txt" contains the following:
# 1,2,3,4
# 5,6,7,8
# 9,10,11,12
matrix = []
with open("csv_file.txt", "r") as f:
full_file = f.read()
# Split into lines.
lines = full_file.strip().split("\n")
for line in lines:
# Split on commas.
elements = line.strip().split(",")
matrix.append([])
# Convert to integers and store in the list.
for e in elements:
matrix[-1].append(int(e))
print(matrix)
```
If, however, we'd prefer to use something a little less `strip()`-y and `split()`-y, Python also has a core `csv` module built-in:
```
import csv
with open("eggs.csv", "w") as csv_file:
file_writer = csv.writer(csv_file)
row1 = ["Sunny-side up", "Over easy", "Scrambled"]
row2 = ["Spam", "Spam", "More spam"]
file_writer.writerow(row1)
file_writer.writerow(row2)
with open("eggs.csv", "r") as csv_file:
print(csv_file.read())
```
Notice that you first create a file reference, just like before. The one added step, though, is passing that reference to the `csv.writer()` function.
Once you've created the `file_writer` object, you can call its `writerow()` function and pass in a list to the function, and it is automatically written to the file in CSV format!
The CSV readers let you do the opposite: read a line of text from a CSV file directly into a list.
```
with open("eggs.csv", "r") as csv_file:
file_reader = csv.reader(csv_file)
for csv_row in file_reader:
print(csv_row)
```
You can use a `for` loop to iterate over the rows in the CSV file. In turn, each row is a list, where each element of the list was separated by a comma.
## Part 2: JavaScript Object Notation (JSON) files
"JSON", short for "JavaScript Object Notation", has emerged as more or less the *de facto* standard format for interacting with online services. Like CSV, it's a text-based format, but is much more flexible than CSV.
Here's an example: an object in JSON format that represents a person.
```
person = """
{"name": "Wes",
"places_lived": ["United States", "Spain", "Germany"],
"pet": null,
"siblings": [{"name": "Scott", "age": 25, "pet": "Zuko"},
{"name": "Katie", "age": 33, "pet": "Cisco"}]
}
"""
```
It looks kind of a like a Python dictionary, doesn't it? You have key-value pairs, and they can accommodate almost any data type. In fact, when JSON objects are converted into native Python data structures, they are represented using dictionaries.
For reading and writing JSON objects, we can use the built-in `json` Python module.
```
import json
```
(*Aside*: with CSV files, it was fairly straightforward to eschew the built-in `csv` module and do it yourself. With JSON, it is **much harder**; in fact, there really isn't a case where it's advisable to roll your own over using the built-in `json` module)
There are two functions of interest: `dumps()` and `loads()`. One of them takes a JSON string and converts it to a native Python object, while the other does the opposite.
First, we'll take our JSON string and convert it into a Python dictionary:
```
python_dict = json.loads(person)
print(python_dict)
```
And if you want to take a Python dictionary and convert it into a JSON string--perhaps you're about to save it to a file, or send it over the network to someone else--we can do that.
```
json_string = json.dumps(python_dict)
print(json_string)
```
At first glance, these two print-outs may look the same, but if you look closely you'll see some differences. Plus, if you tried to index `json_string["name"]` you'd get some very strange errors. `python_dict["name"]`, on the other hand, should nicely return `"Wes"`.
## Part 3: Extensible Markup Language (XML) files
**AVOID AT ALL COSTS**.
...but if you have to interact with XML data (e.g., you're manually parsing a web page!), Python has a built-in `xml` library.
XML is about as general as it gets when it comes to representing data using structured text; you can represent pretty much anything. HTML is an example of XML in practice.
```
<?xml version="1.0" standalone="yes"?>
<conversation>
<greeting>Hello, world!</greeting>
<response>Stop the planet, I want to get off!</response>
</conversation>
```
This is about the simplest excerpt of XML in existence. The basic idea is you have *tags* (delineated by `<` and `>` symbols) that identify where certain fields begin and end.
Each field has an opening tag, with the name of the field in angled brackets: `<field>`. The closing tag is exactly the same, except with a backslash in front of the tag to indicate closing: `</field>`
These tags can also have their own custom *attributes* that slightly tweak their behavior (e.g. the `standalone="yes"` attribute in the opening `<?xml` tag).
You've probably noticed there is a very strong *hierarchy* of terms in XML. This is not unlike JSON in many ways, and for this reason the following piece of advice is the same: don't try to roll your own XML parser. You'll pull out your hair.
The XML file we'll look at comes directly from the [Python documentation for its XML parser](https://docs.python.org/3.5/library/xml.etree.elementtree.html):
```
<?xml version="1.0"?>
<data>
<country name="Liechtenstein">
<rank>1</rank>
<year>2008</year>
<gdppc>141100</gdppc>
<neighbor name="Austria" direction="E"/>
<neighbor name="Switzerland" direction="W"/>
</country>
<country name="Singapore">
<rank>4</rank>
<year>2011</year>
<gdppc>59900</gdppc>
<neighbor name="Malaysia" direction="N"/>
</country>
<country name="Panama">
<rank>68</rank>
<year>2011</year>
<gdppc>13600</gdppc>
<neighbor name="Costa Rica" direction="W"/>
<neighbor name="Colombia" direction="E"/>
</country>
</data>
```
```
import xml.etree.ElementTree as ET # See, even the import statement is stupid complicated.
tree = ET.parse('xml_file.txt')
root = tree.getroot()
print(root.tag) # The root node is "data", so that's what we should see here.
```
With the root node, we have access to all the "child" data beneath it, such as the various country names:
```
for child in root:
print("Tag: \"{}\" :: Name: \"{}\"".format(child.tag, child.attrib["name"]))
```
## Part 4: Binary files
What happens when we're not dealing with *text*? After all, images and videos are most certainly not encoded using text. Furthermore, if memory is an issue, converting text into binary formats can help save space.
There are two primary options for reading and writing binary files.
1. `pickle`, or "pickling", is native in Python and very flexible.
2. NumPy's binary format, which works very well for NumPy arrays but not much else.
Pickle has some similarities with JSON. In particular, it uses the same method names, `dumps()` and `loads()`, for converting between native Python objects and the raw data format. There are several differences, however.
- Most notably, JSON is text-based whereas pickle is binary. You could open up a JSON file and read the text yourself. Not the case with pickled files.
- While JSON is used widely outside of Python, pickle is specific to Python and its objects. Consequently, JSON only works on a subset of Python data structures; pickle, on the other hand, works on just about everything.
Here's an example of saving (or "serializing") a dictionary using pickle instead of JSON:
```
import pickle
# We'll use the `python_dict` object from before.
binary_object = pickle.dumps(python_dict)
print(binary_object)
```
You can kinda see some English in there--mainly, the string constants. But everything else has been encoded in binary. It's much more space-efficient, but complete gibberish until you convert it back into a text format (e.g. JSON) or native Python object (e.g. dictionary).
If, on the other hand, you're using NumPy arrays, then you can use its own built-in binary format for saving and loading your arrays.
```
import numpy as np
# Generate some data and save it.
some_data = np.random.randint(10, size = (3, 3))
print(some_data)
np.save("my_data.npy", some_data)
```
Now we can load it back:
```
my_data = np.load("my_data.npy")
print(my_data)
```
This is by far the easiest format to work with when you're dealing exclusively with NumPy arrays; don't bother with CSV or pickling. You don't even need to set up file descriptors with the NumPy interface.
That said, there are limitations to NumPy serialization: namely, it can only serialize in binary format things that can be stored in NumPy arrays. This does *not* include dictionaries!
`pickle`, on the other hand, can serialize dictionaries (in fact, it specializes in serializing dictionaries), but like NumPy serialization is also not terribly cross-platform capable.
So basically, some core rules of thumb on what binary format to use:
- If it's a NumPy array, use NumPy serialization.
- If it's *not* a NumPy array, but *could* be (e.g. a list of numbers), use NumPy serialization.
- If it's a dictionary, or a structure that mixes string and numeric types, or uses wholesale objects, use `pickle`.
## Review Questions
Some questions to discuss and consider:
1: Dictionaries can be very complex; for a good example, just have a look at how big a dictionary representation of a single Tweet is [https://dev.twitter.com/overview/api/tweets](https://dev.twitter.com/overview/api/tweets): there's `"created_at"`, which is a string indicating the time the tweet was created; `"contributors"`, which is a dictionary unto itself identifying users participating in a thread; `"entities"`, a dictionary of lists that includes hashtags and URLs in the tweet; and `"user"`, which is another gargantuan dictionary containing all the information about the author of the tweet. What would be a good format to store these tweets in on the hard drive? What if we were sending these tweets somewhere, such as a smartphone app; would we use a different format? Explain.
2: You can actually read raw bytes of a binary file using the standard Python `open()` function, provided you supply the special `"b"` flag to indicate a binary format. Can you imagine any circumstances under which you'd read a binary file this way?
3: Is there any other format in which we could store the example XML data from this lecture such that we could avoid using XML entirely?
4: NumPy itself has limited CSV-reading capabilities in `numpy.loadtxt`. Given its limitations in binary serialization as discussed in this lecture, do you imagine there are limitations on what kind of data it can read from CSV files?
5: What kind of format (binary or text) is a .png image? Could it be stored as the other format? How?
## Course Administrivia
- A6 due today!
- Review session #3 next Wednesday (Oct 12) in preparation for the midterm!
- Midterm will be an **in-class, written exam** on Thursday, October 13 (one week from today).
- GII Symposium next Tuesday (Oct 11) at the GA Center. Come and check out the posters and chat with the presenters, then find me for extra credit!
## Additional Resources
1. Matthes, Eric. *Python Crash Course*, Chapter 10. 2016. ISBN-13: 978-1593276034
2. McKinney, Wes. *Python for Data Analysis*, Chapter 6. 2013. ISBN-13: 978-1449319793
|
github_jupyter
|
# File "csv_file.txt" contains the following:
# 1,2,3,4
# 5,6,7,8
# 9,10,11,12
matrix = []
with open("csv_file.txt", "r") as f:
full_file = f.read()
# Split into lines.
lines = full_file.strip().split("\n")
for line in lines:
# Split on commas.
elements = line.strip().split(",")
matrix.append([])
# Convert to integers and store in the list.
for e in elements:
matrix[-1].append(int(e))
print(matrix)
import csv
with open("eggs.csv", "w") as csv_file:
file_writer = csv.writer(csv_file)
row1 = ["Sunny-side up", "Over easy", "Scrambled"]
row2 = ["Spam", "Spam", "More spam"]
file_writer.writerow(row1)
file_writer.writerow(row2)
with open("eggs.csv", "r") as csv_file:
print(csv_file.read())
with open("eggs.csv", "r") as csv_file:
file_reader = csv.reader(csv_file)
for csv_row in file_reader:
print(csv_row)
person = """
{"name": "Wes",
"places_lived": ["United States", "Spain", "Germany"],
"pet": null,
"siblings": [{"name": "Scott", "age": 25, "pet": "Zuko"},
{"name": "Katie", "age": 33, "pet": "Cisco"}]
}
"""
import json
python_dict = json.loads(person)
print(python_dict)
json_string = json.dumps(python_dict)
print(json_string)
<?xml version="1.0" standalone="yes"?>
<conversation>
<greeting>Hello, world!</greeting>
<response>Stop the planet, I want to get off!</response>
</conversation>
<?xml version="1.0"?>
<data>
<country name="Liechtenstein">
<rank>1</rank>
<year>2008</year>
<gdppc>141100</gdppc>
<neighbor name="Austria" direction="E"/>
<neighbor name="Switzerland" direction="W"/>
</country>
<country name="Singapore">
<rank>4</rank>
<year>2011</year>
<gdppc>59900</gdppc>
<neighbor name="Malaysia" direction="N"/>
</country>
<country name="Panama">
<rank>68</rank>
<year>2011</year>
<gdppc>13600</gdppc>
<neighbor name="Costa Rica" direction="W"/>
<neighbor name="Colombia" direction="E"/>
</country>
</data>
import xml.etree.ElementTree as ET # See, even the import statement is stupid complicated.
tree = ET.parse('xml_file.txt')
root = tree.getroot()
print(root.tag) # The root node is "data", so that's what we should see here.
for child in root:
print("Tag: \"{}\" :: Name: \"{}\"".format(child.tag, child.attrib["name"]))
import pickle
# We'll use the `python_dict` object from before.
binary_object = pickle.dumps(python_dict)
print(binary_object)
import numpy as np
# Generate some data and save it.
some_data = np.random.randint(10, size = (3, 3))
print(some_data)
np.save("my_data.npy", some_data)
my_data = np.load("my_data.npy")
print(my_data)
| 0.328637 | 0.959154 |
```
#hide
#skip
! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab
```
# Tabular training
> How to use the tabular application in fastai
To illustrate the tabular application, we will use the example of the [Adult dataset](https://archive.ics.uci.edu/ml/datasets/Adult) where we have to predict if a person is earning more or less than $50k per year using some general data.
```
from fastai.tabular.all import *
```
We can download a sample of this dataset with the usual `untar_data` command:
```
path = untar_data(URLs.ADULT_SAMPLE)
path.ls()
```
Then we can have a look at how the data is structured:
```
df = pd.read_csv(path/'adult.csv')
df.head()
```
Some of the columns are continuous (like age) and we will treat them as float numbers we can feed our model directly. Others are categorical (like workclass or education) and we will convert them to a unique index that we will feed to embedding layers. We can specify our categorical and continuous column names, as well as the name of the dependent variable in `TabularDataLoaders` factory methods:
```
dls = TabularDataLoaders.from_csv(path/'adult.csv', path=path, y_names="salary",
cat_names = ['workclass', 'education', 'marital-status', 'occupation', 'relationship', 'race'],
cont_names = ['age', 'fnlwgt', 'education-num'],
procs = [Categorify, FillMissing, Normalize])
```
The last part is the list of pre-processors we apply to our data:
- `Categorify` is going to take every categorical variable and make a map from integer to unique categories, then replace the values by the corresponding index.
- `FillMissing` will fill the missing values in the continuous variables by the median of existing values (you can choose a specific value if you prefer)
- `Normalize` will normalize the continuous variables (substract the mean and divide by the std)
To further expose what's going on below the surface, let's rewrite this utilizing `fastai`'s `TabularPandas` class. We will need to make one adjustment, which is defining how we want to split our data. By default the factory method above used a random 80/20 split, so we will do the same:
```
splits = RandomSplitter(valid_pct=0.2)(range_of(df))
to = TabularPandas(df, procs=[Categorify, FillMissing,Normalize],
cat_names = ['workclass', 'education', 'marital-status', 'occupation', 'relationship', 'race'],
cont_names = ['age', 'fnlwgt', 'education-num'],
y_names='salary',
splits=splits)
```
Once we build our `TabularPandas` object, our data is completely preprocessed as seen below:
```
to.xs.iloc[:2]
```
Now we can build our `DataLoaders` again:
```
dls = to.dataloaders(bs=64)
```
> Later we will explore why using `TabularPandas` to preprocess will be valuable.
The `show_batch` method works like for every other application:
```
dls.show_batch()
```
We can define a model using the `tabular_learner` method. When we define our model, `fastai` will try to infer the loss function based on our `y_names` earlier.
**Note**: Sometimes with tabular data, your `y`'s may be encoded (such as 0 and 1). In such a case you should explicitly pass `y_block = CategoryBlock` in your constructor so `fastai` won't presume you are doing regression.
```
learn = tabular_learner(dls, metrics=accuracy)
```
And we can train that model with the `fit_one_cycle` method (the `fine_tune` method won't be useful here since we don't have a pretrained model).
```
learn.fit_one_cycle(1)
```
We can then have a look at some predictions:
```
learn.show_results()
```
Or use the predict method on a row:
```
row, clas, probs = learn.predict(df.iloc[0])
row.show()
clas, probs
```
To get prediction on a new dataframe, you can use the `test_dl` method of the `DataLoaders`. That dataframe does not need to have the dependent variable in its column.
```
test_df = df.copy()
test_df.drop(['salary'], axis=1, inplace=True)
dl = learn.dls.test_dl(test_df)
```
Then `Learner.get_preds` will give you the predictions:
```
learn.get_preds(dl=dl)
```
> Note: Since machine learning models can't magically understand categories it was never trained on, the data should reflect this. If there are different missing values in your test data you should address this before training
## `fastai` with Other Libraries
As mentioned earlier, `TabularPandas` is a powerful and easy preprocessing tool for tabular data. Integration with libraries such as Random Forests and XGBoost requires only one extra step, that the `.dataloaders` call did for us. Let's look at our `to` again. It's values are stored in a `DataFrame` like object, where we can extract the `cats`, `conts,` `xs` and `ys` if we want to:
```
to.xs[:3]
```
Now that everything is encoded, you can then send this off to XGBoost or Random Forests by extracting the train and validation sets and their values:
```
X_train, y_train = to.train.xs, to.train.ys.values.ravel()
X_test, y_test = to.valid.xs, to.valid.ys.values.ravel()
```
And now we can directly send this in!
|
github_jupyter
|
#hide
#skip
! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab
from fastai.tabular.all import *
path = untar_data(URLs.ADULT_SAMPLE)
path.ls()
df = pd.read_csv(path/'adult.csv')
df.head()
dls = TabularDataLoaders.from_csv(path/'adult.csv', path=path, y_names="salary",
cat_names = ['workclass', 'education', 'marital-status', 'occupation', 'relationship', 'race'],
cont_names = ['age', 'fnlwgt', 'education-num'],
procs = [Categorify, FillMissing, Normalize])
splits = RandomSplitter(valid_pct=0.2)(range_of(df))
to = TabularPandas(df, procs=[Categorify, FillMissing,Normalize],
cat_names = ['workclass', 'education', 'marital-status', 'occupation', 'relationship', 'race'],
cont_names = ['age', 'fnlwgt', 'education-num'],
y_names='salary',
splits=splits)
to.xs.iloc[:2]
dls = to.dataloaders(bs=64)
dls.show_batch()
learn = tabular_learner(dls, metrics=accuracy)
learn.fit_one_cycle(1)
learn.show_results()
row, clas, probs = learn.predict(df.iloc[0])
row.show()
clas, probs
test_df = df.copy()
test_df.drop(['salary'], axis=1, inplace=True)
dl = learn.dls.test_dl(test_df)
learn.get_preds(dl=dl)
to.xs[:3]
X_train, y_train = to.train.xs, to.train.ys.values.ravel()
X_test, y_test = to.valid.xs, to.valid.ys.values.ravel()
| 0.339828 | 0.988917 |
# Unify and clean-up intersections of divided roads
Divided roads are represented by separate centerline edges. The intersection of two divided roads thus creates 4 nodes, representing where each edge intersects a perpendicular edge. These 4 nodes represent a single intersection in the real world. Roundabouts similarly create a cluster of intersections where each edge connects to the roundabout. This function cleans up these clusters by buffering their points to an arbitrary distance, merging overlapping buffers, and returning a GeoSeries of their centroids. For best results, the tolerance argument should be adjusted to approximately match street design standards in the specific street network.
- [Overview of OSMnx](http://geoffboeing.com/2016/11/osmnx-python-street-networks/)
- [GitHub repo](https://github.com/gboeing/osmnx)
- [Examples, demos, tutorials](https://github.com/gboeing/osmnx-examples)
- [Documentation](https://osmnx.readthedocs.io/en/stable/)
- [Journal article/citation](http://geoffboeing.com/publications/osmnx-complex-street-networks/)
```
import osmnx as ox, matplotlib.pyplot as plt, numpy as np
ox.config(use_cache=True, log_console=True)
%matplotlib inline
# get a street network and plot it with all edge intersections
address = '2700 Shattuck Ave, Berkeley, CA'
G = ox.graph_from_address(address, network_type='drive', distance=750)
G_proj = ox.project_graph(G)
fig, ax = ox.plot_graph(G_proj, fig_height=10, node_color='orange', node_size=30,
node_zorder=2, node_edgecolor='k')
```
### Clean up the intersections
We'll specify that any nodes with 15 meters of each other in this network are part of the same intersection. Adjust this tolerance based on the street design standards in the community you are examining, and use a projected graph to work in meaningful units like meters. We'll also specify that we do not want dead-ends returned in our list of cleaned intersections. Then we extract their xy coordinates and plot it to show how the clean intersections below compare to the topological edge intersections above.
```
# clean up the intersections and extract their xy coords
intersections = ox.clean_intersections(G_proj, tolerance=15, dead_ends=False)
points = np.array([point.xy for point in intersections])
# plot the cleaned-up intersections
fig, ax = ox.plot_graph(G_proj, fig_height=10, show=False, close=False, node_alpha=0)
ax.scatter(x=points[:,0], y=points[:,1], zorder=2, color='#66ccff', edgecolors='k')
plt.show()
```
Note that these cleaned up intersections give us more accurate intersection counts and densities, but do not alter or integrate with the network's topology.
|
github_jupyter
|
import osmnx as ox, matplotlib.pyplot as plt, numpy as np
ox.config(use_cache=True, log_console=True)
%matplotlib inline
# get a street network and plot it with all edge intersections
address = '2700 Shattuck Ave, Berkeley, CA'
G = ox.graph_from_address(address, network_type='drive', distance=750)
G_proj = ox.project_graph(G)
fig, ax = ox.plot_graph(G_proj, fig_height=10, node_color='orange', node_size=30,
node_zorder=2, node_edgecolor='k')
# clean up the intersections and extract their xy coords
intersections = ox.clean_intersections(G_proj, tolerance=15, dead_ends=False)
points = np.array([point.xy for point in intersections])
# plot the cleaned-up intersections
fig, ax = ox.plot_graph(G_proj, fig_height=10, show=False, close=False, node_alpha=0)
ax.scatter(x=points[:,0], y=points[:,1], zorder=2, color='#66ccff', edgecolors='k')
plt.show()
| 0.536313 | 0.994066 |
#Mount Google Drive
```
import sys
import os
from google.colab import drive
drive.mount('/content/gdrive')
# Change working directory to be current folder
import os
os.chdir('/content/gdrive/My Drive/iss/babydetect/')
```
## Environment setup
```
!pip install tensorflow.io
!pip install ffmpeg moviepy
!pip install librosa
!apt install libasound2-dev portaudio19-dev libportaudio2 libportaudiocpp0 ffmpeg
!pip install PyAudio
```
# Sound classification with YAMNet
YAMNet is a deep net that predicts 521 audio event [classes](https://github.com/tensorflow/models/blob/master/research/audioset/yamnet/yamnet_class_map.csv) from the [AudioSet-YouTube corpus](http://g.co/audioset) it was trained on. It employs the
[Mobilenet_v1](https://arxiv.org/pdf/1704.04861.pdf) depthwise-separable
convolution architecture.
```
import librosa
import soundfile as sf
import tensorflow as tf
import tensorflow_datasets as tfds
import tensorflow_hub as hub
import tensorflow_io as tfio
import csv
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from IPython import display
import moviepy.editor as mp
from scipy.io import wavfile
from scipy.signal import resample
```
Load the Model from TensorFlow Hub.
Note: to read the documentation just follow the model's [url](https://tfhub.dev/google/yamnet/1)
```
# Load the model.
yamnet_model = hub.load('YAMNet')
```
The labels file will be loaded from the models assets and is present at `model.class_map_path()`.
You will load it on the `class_names` variable.
```
# solution: loading label names
class_map_path = yamnet_model.class_map_path().numpy().decode('utf-8')
class_names =list(pd.read_csv(class_map_path)['display_name'])
for name in class_names[:5]:
print(name)
```
Add a method to convert a loaded audio is on the proper sample_rate (16K), otherwise it would affect the model's results.
Returned wav_data has been normalized to values in [-1.0, 1.0] (as stated in the model's documentation).
```
@tf.function
def load_wav_16k_mono(filename):
""" read in a waveform file and convert to 16 kHz mono """
file_contents = tf.io.read_file(filename)
wav, sample_rate = tf.audio.decode_wav(file_contents,
desired_channels=1)
wav = tf.squeeze(wav, axis=-1)
sample_rate = tf.cast(sample_rate, dtype=tf.int64)
wav = tfio.audio.resample(wav, rate_in=sample_rate, rate_out=16000)
return wav
```
## Preparing the sound file
The audio file should be a mono wav file at 16kHz sample rate.
```
wav_file_name = './datasets/ESC-50-master/audio/1-187207-A-20.wav'
wav_data = load_wav_16k_mono(wav_file_name)
# Play the audio file.
display.Audio(wav_data, rate=16000)
plt.plot(wav_data)
```
## Executing the Model
Now the easy part: using the data already prepared, you just call the model and get the: scores, embedding and the spectrogram.
The score is the main result you will use.
The spectrogram you will use to do some visualizations later.
```
# Run the model, check the output.
scores, embeddings, spectrogram = yamnet_model(wav_data)
scores_np = scores.numpy()
spectrogram_np = spectrogram.numpy()
infered_class = class_names[scores_np.mean(axis=0).argmax()]
print(f'The main sound is: {infered_class}')
class_scores = tf.reduce_mean(scores, axis=0)
top_class = tf.argmax(class_scores)
infered_class = class_names[top_class]
print(f'The main sound is: {infered_class}')
print(f'The embeddings shape: {embeddings.shape}')
```
## Visualization
YAMNet also returns some additional information that we can use for visualization.
Let's take a look on the Waveform, spectrogram and the top classes inferred.
```
plt.figure(figsize=(10, 6))
# Plot the waveform.
plt.subplot(3, 1, 1)
plt.plot(wav_data)
plt.xlim([0, len(wav_data)])
# Plot the log-mel spectrogram (returned by the model).
plt.subplot(3, 1, 2)
plt.imshow(spectrogram_np.T, aspect='auto', interpolation='nearest', origin='lower')
# Plot and label the model output scores for the top-scoring classes.
mean_scores = np.mean(scores, axis=0)
top_n = 10
top_class_indices = np.argsort(mean_scores)[::-1][:top_n]
plt.subplot(3, 1, 3)
plt.imshow(scores_np[:, top_class_indices].T, aspect='auto', interpolation='nearest', cmap='gray_r')
# patch_padding = (PATCH_WINDOW_SECONDS / 2) / PATCH_HOP_SECONDS
# values from the model documentation
patch_padding = (0.025 / 2) / 0.01
plt.xlim([-patch_padding-0.5, scores.shape[0] + patch_padding-0.5])
# Label the top_N classes.
yticks = range(0, top_n, 1)
plt.yticks(yticks, [class_names[top_class_indices[x]] for x in yticks])
_ = plt.ylim(-0.5 + np.array([top_n, 0]))
```
## ESC-50 dataset
The ESC-50 dataset, well described here, is a labeled collection of 2000 environmental audio recordings (each 5 seconds long). The data consists of 50 classes, with 40 examples per class
```
_ = tf.keras.utils.get_file('esc-50.zip',
'https://github.com/karoldvl/ESC-50/archive/master.zip',
cache_dir='./',
cache_subdir='datasets',
extract=True)
```
## Explore the data
```
esc50_csv = './datasets/ESC-50-master/meta/esc50.csv'
base_data_path = './datasets/ESC-50-master/audio/'
pd_data = pd.read_csv(esc50_csv)
pd_data.head()
```
## Filter the data
```
my_classes = ['crying_baby', 'laughing']
saved_model_path = './baby_crying_yamnet1'
filtered_pd_crying = pd_data[pd_data.category.isin(['crying_baby'])]
print(len(filtered_pd_crying))
map_class_to_id = {'crying_baby':0, 'laughing':1}
filtered_pd = pd_data[pd_data.category.isin(my_classes)]
class_id = filtered_pd['category'].apply(lambda name: map_class_to_id[name])
filtered_pd = filtered_pd.assign(target=class_id)
full_path = filtered_pd['filename'].apply(lambda row: os.path.join(base_data_path, row))
filtered_pd = filtered_pd.assign(filename=full_path)
filtered_pd.head(10)
```
## Load the audio files and retrieve embeddings
```
filenames = filtered_pd['filename']
targets = filtered_pd['target']
folds = filtered_pd['fold']
main_ds = tf.data.Dataset.from_tensor_slices((filenames, targets, folds))
main_ds.element_spec
def load_wav_for_map(filename, label, fold):
return load_wav_16k_mono(filename), label, fold
#main_ds = main_ds.map(lambda a,b,c: tf.py_function(load_wav_for_map, [a, b, c], [tf.float32,tf.int64,tf.int64]))
main_ds = main_ds.map(load_wav_for_map)
main_ds.element_spec
def extract_embedding(wav_data, label, fold):
''' run YAMNet to extract embedding from the wav data '''
scores, embeddings, spectrogram = yamnet_model(wav_data)
num_embeddings = tf.shape(embeddings)[0]
return (embeddings,
tf.repeat(label, num_embeddings),
tf.repeat(fold, num_embeddings))
# extract embedding
main_ds = main_ds.map(extract_embedding).unbatch()
main_ds.element_spec
cached_ds = main_ds.cache()
train_ds = cached_ds.filter(lambda embedding, label, fold: fold < 4)
val_ds = cached_ds.filter(lambda embedding, label, fold: fold == 4)
test_ds = cached_ds.filter(lambda embedding, label, fold: fold == 5)
# remove the folds column now that it's not needed anymore
remove_fold_column = lambda embedding, label, fold: (embedding, label)
train_ds = train_ds.map(remove_fold_column)
val_ds = val_ds.map(remove_fold_column)
test_ds = test_ds.map(remove_fold_column)
train_ds = train_ds.cache().shuffle(1000).batch(32).prefetch(tf.data.AUTOTUNE)
val_ds = val_ds.cache().batch(32).prefetch(tf.data.AUTOTUNE)
test_ds = test_ds.cache().batch(32).prefetch(tf.data.AUTOTUNE)
print(train_ds)
```
## Create new model
```
new_model = tf.keras.Sequential([
tf.keras.layers.Input(shape=(1024),
dtype=tf.float32,
name='input_embedding'),
tf.keras.layers.Dense(512, activation='relu'),
tf.keras.layers.Dense(len(my_classes))
], name='new_model')
new_model.summary()
new_model.compile(loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
optimizer="adam",
metrics=['accuracy'])
callback = tf.keras.callbacks.EarlyStopping(monitor='loss',
patience=3,
restore_best_weights=True)
history = new_model.fit(train_ds,
epochs=20,
validation_data=val_ds,
callbacks=callback)
```
Lets run the evaluate method on the test data just to be sure there's no overfitting.
```
loss, accuracy = new_model.evaluate(test_ds)
print("Loss: ", loss)
print("Accuracy: ", accuracy)
```
## Test your model
```
test_laughing_data = load_wav_16k_mono('./datasets/ESC-50-master/audio/4-155670-A-26.wav')
scores, embeddings, spectrogram = yamnet_model(test_laughing_data)
result = new_model(embeddings).numpy()
print(result)
score = result.mean(axis=0).argmax()
threshold = False
for i, j in result:
if i > 5.0:
threshold = True
break
if threshold == False:
score = 1
infered_class = my_classes[score]
print(f'The main sound is: {infered_class}')
```
## Save a model that can directly take a wav file as input
set threshold 5.0 for max score
```
class ThresholdMeanLayer(tf.keras.layers.Layer):
def __init__(self, axis=0, **kwargs):
super(ThresholdMeanLayer, self).__init__(**kwargs)
self.axis = axis
def call(self, input):
mean_scores = tf.math.reduce_mean(input, axis=self.axis)
max_score = tf.argmax(mean_scores)
return tf.cond(tf.reduce_max(input[:,0], axis=0) > 5, lambda:max_score, lambda:tf.cast(1, tf.int64))
input_segment = tf.keras.layers.Input(shape=(), dtype=tf.float32, name='audio')
embedding_extraction_layer = hub.KerasLayer('YAMNet',
trainable=False,
name='yamnet')
_, embeddings_output, _ = embedding_extraction_layer(input_segment)
serving_outputs = new_model(embeddings_output)
serving_outputs = ThresholdMeanLayer(axis=0, name='classifier')(serving_outputs)
serving_model = tf.keras.Model(input_segment, serving_outputs)
serving_model.save(saved_model_path, include_optimizer=False)
tf.keras.utils.plot_model(serving_model)
```
## Test new model
## Loading video
```
#my_clip = mp.VideoFileClip(r"./datasets/Babies_Crying.mp4")
#my_clip.audio.write_audiofile(r"./datasets/Babies_Crying.wav")
my_clip = mp.VideoFileClip(r"./datasets/climbing.mp4")
my_clip.audio.write_audiofile(r"./datasets/climbing.wav")
#test_laughing_data = load_wav_16k_mono('./datasets/ESC-50-master/audio/4-155670-A-26.wav')
#test_crying_data = load_wav_16k_mono('./datasets/ESC-50-master/audio/4-167077-A-20.wav')
#test_data = load_wav_16k_mono('./datasets/Babies_Crying.wav')
test_data = load_wav_16k_mono('./datasets/climbing.wav')
# loading new model
reloaded_model = tf.saved_model.load(saved_model_path)
# test in new data file
reloaded_results = reloaded_model(test_data)
print(reloaded_results)
baby_sound = my_classes[reloaded_results]
print(f'The main sound is: {baby_sound}')
```
## Read audio file
```
sample_rate = 16000
rate = 44100
duration = len(test_data)/sample_rate
print(f'Total duration: {duration:.2f}s')
for i in range(0, int(duration), 5):
start = i*sample_rate
end = (i+5)*sample_rate
print('duration from {:d} -- {:d}'.format(i, i+5))
wav_data = test_data[start:end]
reloaded_results = reloaded_model(wav_data)
baby_sound = my_classes[reloaded_results]
print(f'The main sound is: {baby_sound}, score: {reloaded_results}, max: {reloaded_results}')
filename = 'clip-{:d}.wav'.format(i)
data = np.random.uniform(-1, 1, size=(rate * 10, 2))
sf.write(filename, wav_data, sample_rate, subtype='PCM_24')
```
## Real-Time audio
```
import pyaudio
p = pyaudio.PyAudio()
print(p.get_device_count())
FORMAT = pyaudio.paInt16
CHANNELS = 1
RATE = 44100
RECORD_SECONDS = 5
CHUNK = int(RATE/20)
stream = p.open(format=FORMAT,
channels=CHANNELS,
rate=RATE,
input = True,
frames_per_buffer=CHUNK)
while True:
frames = []
for _ in range(0, int(RATE / CHUNK * RECORD_SECONDS)):
data = stream.read(CHUNK, exception_on_overflow=False)
frames.append(np.fromstring(data, dtype=np.float32))
npdata = np.hstack(frames)
wav_data = AudioClip.from_np(npdata, RATE)
#check using model
reloaded_results = reloaded_model(wav_data)
baby_sound = my_classes[tf.argmax(reloaded_results)]
print(f'The main sound is: {baby_sound}')
```
|
github_jupyter
|
import sys
import os
from google.colab import drive
drive.mount('/content/gdrive')
# Change working directory to be current folder
import os
os.chdir('/content/gdrive/My Drive/iss/babydetect/')
!pip install tensorflow.io
!pip install ffmpeg moviepy
!pip install librosa
!apt install libasound2-dev portaudio19-dev libportaudio2 libportaudiocpp0 ffmpeg
!pip install PyAudio
import librosa
import soundfile as sf
import tensorflow as tf
import tensorflow_datasets as tfds
import tensorflow_hub as hub
import tensorflow_io as tfio
import csv
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from IPython import display
import moviepy.editor as mp
from scipy.io import wavfile
from scipy.signal import resample
# Load the model.
yamnet_model = hub.load('YAMNet')
# solution: loading label names
class_map_path = yamnet_model.class_map_path().numpy().decode('utf-8')
class_names =list(pd.read_csv(class_map_path)['display_name'])
for name in class_names[:5]:
print(name)
@tf.function
def load_wav_16k_mono(filename):
""" read in a waveform file and convert to 16 kHz mono """
file_contents = tf.io.read_file(filename)
wav, sample_rate = tf.audio.decode_wav(file_contents,
desired_channels=1)
wav = tf.squeeze(wav, axis=-1)
sample_rate = tf.cast(sample_rate, dtype=tf.int64)
wav = tfio.audio.resample(wav, rate_in=sample_rate, rate_out=16000)
return wav
wav_file_name = './datasets/ESC-50-master/audio/1-187207-A-20.wav'
wav_data = load_wav_16k_mono(wav_file_name)
# Play the audio file.
display.Audio(wav_data, rate=16000)
plt.plot(wav_data)
# Run the model, check the output.
scores, embeddings, spectrogram = yamnet_model(wav_data)
scores_np = scores.numpy()
spectrogram_np = spectrogram.numpy()
infered_class = class_names[scores_np.mean(axis=0).argmax()]
print(f'The main sound is: {infered_class}')
class_scores = tf.reduce_mean(scores, axis=0)
top_class = tf.argmax(class_scores)
infered_class = class_names[top_class]
print(f'The main sound is: {infered_class}')
print(f'The embeddings shape: {embeddings.shape}')
plt.figure(figsize=(10, 6))
# Plot the waveform.
plt.subplot(3, 1, 1)
plt.plot(wav_data)
plt.xlim([0, len(wav_data)])
# Plot the log-mel spectrogram (returned by the model).
plt.subplot(3, 1, 2)
plt.imshow(spectrogram_np.T, aspect='auto', interpolation='nearest', origin='lower')
# Plot and label the model output scores for the top-scoring classes.
mean_scores = np.mean(scores, axis=0)
top_n = 10
top_class_indices = np.argsort(mean_scores)[::-1][:top_n]
plt.subplot(3, 1, 3)
plt.imshow(scores_np[:, top_class_indices].T, aspect='auto', interpolation='nearest', cmap='gray_r')
# patch_padding = (PATCH_WINDOW_SECONDS / 2) / PATCH_HOP_SECONDS
# values from the model documentation
patch_padding = (0.025 / 2) / 0.01
plt.xlim([-patch_padding-0.5, scores.shape[0] + patch_padding-0.5])
# Label the top_N classes.
yticks = range(0, top_n, 1)
plt.yticks(yticks, [class_names[top_class_indices[x]] for x in yticks])
_ = plt.ylim(-0.5 + np.array([top_n, 0]))
_ = tf.keras.utils.get_file('esc-50.zip',
'https://github.com/karoldvl/ESC-50/archive/master.zip',
cache_dir='./',
cache_subdir='datasets',
extract=True)
esc50_csv = './datasets/ESC-50-master/meta/esc50.csv'
base_data_path = './datasets/ESC-50-master/audio/'
pd_data = pd.read_csv(esc50_csv)
pd_data.head()
my_classes = ['crying_baby', 'laughing']
saved_model_path = './baby_crying_yamnet1'
filtered_pd_crying = pd_data[pd_data.category.isin(['crying_baby'])]
print(len(filtered_pd_crying))
map_class_to_id = {'crying_baby':0, 'laughing':1}
filtered_pd = pd_data[pd_data.category.isin(my_classes)]
class_id = filtered_pd['category'].apply(lambda name: map_class_to_id[name])
filtered_pd = filtered_pd.assign(target=class_id)
full_path = filtered_pd['filename'].apply(lambda row: os.path.join(base_data_path, row))
filtered_pd = filtered_pd.assign(filename=full_path)
filtered_pd.head(10)
filenames = filtered_pd['filename']
targets = filtered_pd['target']
folds = filtered_pd['fold']
main_ds = tf.data.Dataset.from_tensor_slices((filenames, targets, folds))
main_ds.element_spec
def load_wav_for_map(filename, label, fold):
return load_wav_16k_mono(filename), label, fold
#main_ds = main_ds.map(lambda a,b,c: tf.py_function(load_wav_for_map, [a, b, c], [tf.float32,tf.int64,tf.int64]))
main_ds = main_ds.map(load_wav_for_map)
main_ds.element_spec
def extract_embedding(wav_data, label, fold):
''' run YAMNet to extract embedding from the wav data '''
scores, embeddings, spectrogram = yamnet_model(wav_data)
num_embeddings = tf.shape(embeddings)[0]
return (embeddings,
tf.repeat(label, num_embeddings),
tf.repeat(fold, num_embeddings))
# extract embedding
main_ds = main_ds.map(extract_embedding).unbatch()
main_ds.element_spec
cached_ds = main_ds.cache()
train_ds = cached_ds.filter(lambda embedding, label, fold: fold < 4)
val_ds = cached_ds.filter(lambda embedding, label, fold: fold == 4)
test_ds = cached_ds.filter(lambda embedding, label, fold: fold == 5)
# remove the folds column now that it's not needed anymore
remove_fold_column = lambda embedding, label, fold: (embedding, label)
train_ds = train_ds.map(remove_fold_column)
val_ds = val_ds.map(remove_fold_column)
test_ds = test_ds.map(remove_fold_column)
train_ds = train_ds.cache().shuffle(1000).batch(32).prefetch(tf.data.AUTOTUNE)
val_ds = val_ds.cache().batch(32).prefetch(tf.data.AUTOTUNE)
test_ds = test_ds.cache().batch(32).prefetch(tf.data.AUTOTUNE)
print(train_ds)
new_model = tf.keras.Sequential([
tf.keras.layers.Input(shape=(1024),
dtype=tf.float32,
name='input_embedding'),
tf.keras.layers.Dense(512, activation='relu'),
tf.keras.layers.Dense(len(my_classes))
], name='new_model')
new_model.summary()
new_model.compile(loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
optimizer="adam",
metrics=['accuracy'])
callback = tf.keras.callbacks.EarlyStopping(monitor='loss',
patience=3,
restore_best_weights=True)
history = new_model.fit(train_ds,
epochs=20,
validation_data=val_ds,
callbacks=callback)
loss, accuracy = new_model.evaluate(test_ds)
print("Loss: ", loss)
print("Accuracy: ", accuracy)
test_laughing_data = load_wav_16k_mono('./datasets/ESC-50-master/audio/4-155670-A-26.wav')
scores, embeddings, spectrogram = yamnet_model(test_laughing_data)
result = new_model(embeddings).numpy()
print(result)
score = result.mean(axis=0).argmax()
threshold = False
for i, j in result:
if i > 5.0:
threshold = True
break
if threshold == False:
score = 1
infered_class = my_classes[score]
print(f'The main sound is: {infered_class}')
class ThresholdMeanLayer(tf.keras.layers.Layer):
def __init__(self, axis=0, **kwargs):
super(ThresholdMeanLayer, self).__init__(**kwargs)
self.axis = axis
def call(self, input):
mean_scores = tf.math.reduce_mean(input, axis=self.axis)
max_score = tf.argmax(mean_scores)
return tf.cond(tf.reduce_max(input[:,0], axis=0) > 5, lambda:max_score, lambda:tf.cast(1, tf.int64))
input_segment = tf.keras.layers.Input(shape=(), dtype=tf.float32, name='audio')
embedding_extraction_layer = hub.KerasLayer('YAMNet',
trainable=False,
name='yamnet')
_, embeddings_output, _ = embedding_extraction_layer(input_segment)
serving_outputs = new_model(embeddings_output)
serving_outputs = ThresholdMeanLayer(axis=0, name='classifier')(serving_outputs)
serving_model = tf.keras.Model(input_segment, serving_outputs)
serving_model.save(saved_model_path, include_optimizer=False)
tf.keras.utils.plot_model(serving_model)
#my_clip = mp.VideoFileClip(r"./datasets/Babies_Crying.mp4")
#my_clip.audio.write_audiofile(r"./datasets/Babies_Crying.wav")
my_clip = mp.VideoFileClip(r"./datasets/climbing.mp4")
my_clip.audio.write_audiofile(r"./datasets/climbing.wav")
#test_laughing_data = load_wav_16k_mono('./datasets/ESC-50-master/audio/4-155670-A-26.wav')
#test_crying_data = load_wav_16k_mono('./datasets/ESC-50-master/audio/4-167077-A-20.wav')
#test_data = load_wav_16k_mono('./datasets/Babies_Crying.wav')
test_data = load_wav_16k_mono('./datasets/climbing.wav')
# loading new model
reloaded_model = tf.saved_model.load(saved_model_path)
# test in new data file
reloaded_results = reloaded_model(test_data)
print(reloaded_results)
baby_sound = my_classes[reloaded_results]
print(f'The main sound is: {baby_sound}')
sample_rate = 16000
rate = 44100
duration = len(test_data)/sample_rate
print(f'Total duration: {duration:.2f}s')
for i in range(0, int(duration), 5):
start = i*sample_rate
end = (i+5)*sample_rate
print('duration from {:d} -- {:d}'.format(i, i+5))
wav_data = test_data[start:end]
reloaded_results = reloaded_model(wav_data)
baby_sound = my_classes[reloaded_results]
print(f'The main sound is: {baby_sound}, score: {reloaded_results}, max: {reloaded_results}')
filename = 'clip-{:d}.wav'.format(i)
data = np.random.uniform(-1, 1, size=(rate * 10, 2))
sf.write(filename, wav_data, sample_rate, subtype='PCM_24')
import pyaudio
p = pyaudio.PyAudio()
print(p.get_device_count())
FORMAT = pyaudio.paInt16
CHANNELS = 1
RATE = 44100
RECORD_SECONDS = 5
CHUNK = int(RATE/20)
stream = p.open(format=FORMAT,
channels=CHANNELS,
rate=RATE,
input = True,
frames_per_buffer=CHUNK)
while True:
frames = []
for _ in range(0, int(RATE / CHUNK * RECORD_SECONDS)):
data = stream.read(CHUNK, exception_on_overflow=False)
frames.append(np.fromstring(data, dtype=np.float32))
npdata = np.hstack(frames)
wav_data = AudioClip.from_np(npdata, RATE)
#check using model
reloaded_results = reloaded_model(wav_data)
baby_sound = my_classes[tf.argmax(reloaded_results)]
print(f'The main sound is: {baby_sound}')
| 0.670716 | 0.778607 |
<div align="center">
<h1><img width="30" src="https://madewithml.com/static/images/rounded_logo.png"> <a href="https://madewithml.com/">Made With ML</a></h1>
Applied ML · MLOps · Production
<br>
Join 20K+ developers in learning how to responsibly <a href="https://madewithml.com/about/">deliver value</a> with ML.
</div>
<br>
<div align="center">
<a target="_blank" href="https://newsletter.madewithml.com"><img src="https://img.shields.io/badge/Subscribe-20K-brightgreen"></a>
<a target="_blank" href="https://github.com/GokuMohandas/MadeWithML"><img src="https://img.shields.io/github/stars/GokuMohandas/MadeWithML.svg?style=social&label=Star"></a>
<a target="_blank" href="https://www.linkedin.com/in/goku"><img src="https://img.shields.io/badge/style--5eba00.svg?label=LinkedIn&logo=linkedin&style=social"></a>
<a target="_blank" href="https://twitter.com/GokuMohandas"><img src="https://img.shields.io/twitter/follow/GokuMohandas.svg?label=Follow&style=social"></a>
<p>🔥 Among the <a href="https://github.com/topics/deep-learning" target="_blank">top ML</a> repositories on GitHub</p>
</div>
<br>
<hr>
# Convolutional Neural Networks (CNN)
In this lesson we will explore the basics of Convolutional Neural Networks (CNNs) applied to text for natural language processing (NLP) tasks.
<div align="left">
<a target="_blank" href="https://madewithml.com/courses/ml-foundations/convolutional-neural-networks/"><img src="https://img.shields.io/badge/📖 Read-blog post-9cf"></a>
<a href="https://github.com/GokuMohandas/MadeWithML/blob/main/notebooks/11_Convolutional_Neural_Networks.ipynb" role="button"><img src="https://img.shields.io/static/v1?label=&message=View%20On%20GitHub&color=586069&logo=github&labelColor=2f363d"></a>
<a href="https://colab.research.google.com/github/GokuMohandas/MadeWithML/blob/main/notebooks/11_Convolutional_Neural_Networks.ipynb"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"></a>
</div>
# Overview
At the core of CNNs are filters (aka weights, kernels, etc.) which convolve (slide) across our input to extract relevant features. The filters are initialized randomly but learn to act as feature extractors via parameter sharing.
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/cnn/convolution.gif" width="500">
</div>
* **Objective:** Extract meaningful spatial substructure from encoded data.
* **Advantages:**
* Small number of weights (shared)
* Parallelizable
* Detects spatial substrcutures (feature extractors)
* [Interpretability](https://arxiv.org/abs/1312.6034) via filters
* Can be used for processing in images, text, time-series, etc.
* **Disadvantages:**
* Many hyperparameters (kernel size, strides, etc.) to tune.
* **Miscellaneous:**
* Lot's of deep CNN architectures constantly updated for SOTA performance.
* Very popular feature extractor that's usually prepended onto other architectures.
# Set up
```
import numpy as np
import pandas as pd
import random
import torch
import torch.nn as nn
SEED = 1234
def set_seeds(seed=1234):
"""Set seeds for reproducibility."""
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed) # multi-GPU
# Set seeds for reproducibility
set_seeds(seed=SEED)
# Set device
cuda = True
device = torch.device('cuda' if (
torch.cuda.is_available() and cuda) else 'cpu')
torch.set_default_tensor_type('torch.FloatTensor')
if device.type == 'cuda':
torch.set_default_tensor_type('torch.cuda.FloatTensor')
print (device)
```
## Load data
We will download the [AG News dataset](http://www.di.unipi.it/~gulli/AG_corpus_of_news_articles.html), which consists of 120K text samples from 4 unique classes (`Business`, `Sci/Tech`, `Sports`, `World`)
```
# Load data
url = "https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/datasets/news.csv"
df = pd.read_csv(url, header=0) # load
df = df.sample(frac=1).reset_index(drop=True) # shuffle
df.head()
```
## Preprocessing
We're going to clean up our input data first by doing operations such as lower text, removing stop (filler) words, filters using regular expressions, etc.
```
import nltk
from nltk.corpus import stopwords
from nltk.stem import PorterStemmer
import re
nltk.download('stopwords')
STOPWORDS = stopwords.words('english')
print (STOPWORDS[:5])
porter = PorterStemmer()
def preprocess(text, stopwords=STOPWORDS):
"""Conditional preprocessing on our text unique to our task."""
# Lower
text = text.lower()
# Remove stopwords
pattern = re.compile(r'\b(' + r'|'.join(stopwords) + r')\b\s*')
text = pattern.sub('', text)
# Remove words in paranthesis
text = re.sub(r'\([^)]*\)', '', text)
# Spacing and filters
text = re.sub(r"([-;;.,!?<=>])", r" \1 ", text)
text = re.sub('[^A-Za-z0-9]+', ' ', text) # remove non alphanumeric chars
text = re.sub(' +', ' ', text) # remove multiple spaces
text = text.strip()
return text
# Sample
text = "Great week for the NYSE!"
preprocess(text=text)
# Apply to dataframe
preprocessed_df = df.copy()
preprocessed_df.title = preprocessed_df.title.apply(preprocess)
print (f"{df.title.values[0]}\n\n{preprocessed_df.title.values[0]}")
```
> If you have preprocessing steps like standardization, etc. that are calculated, you need to separate the training and test set first before applying those operations. This is because we cannot apply any knowledge gained from the test set accidentally (data leak) during preprocessing/training. However for global preprocessing steps like the function above where we aren't learning anything from the data itself, we can perform before splitting the data.
## Split data
```
import collections
from sklearn.model_selection import train_test_split
TRAIN_SIZE = 0.7
VAL_SIZE = 0.15
TEST_SIZE = 0.15
def train_val_test_split(X, y, train_size):
"""Split dataset into data splits."""
X_train, X_, y_train, y_ = train_test_split(X, y, train_size=TRAIN_SIZE, stratify=y)
X_val, X_test, y_val, y_test = train_test_split(X_, y_, train_size=0.5, stratify=y_)
return X_train, X_val, X_test, y_train, y_val, y_test
# Data
X = preprocessed_df["title"].values
y = preprocessed_df["category"].values
# Create data splits
X_train, X_val, X_test, y_train, y_val, y_test = train_val_test_split(
X=X, y=y, train_size=TRAIN_SIZE)
print (f"X_train: {X_train.shape}, y_train: {y_train.shape}")
print (f"X_val: {X_val.shape}, y_val: {y_val.shape}")
print (f"X_test: {X_test.shape}, y_test: {y_test.shape}")
print (f"Sample point: {X_train[0]} → {y_train[0]}")
```
## LabelEncoder
Next we'll define a `LabelEncoder` to encode our text labels into unique indices
```
import itertools
class LabelEncoder(object):
"""Label encoder for tag labels."""
def __init__(self, class_to_index={}):
self.class_to_index = class_to_index
self.index_to_class = {v: k for k, v in self.class_to_index.items()}
self.classes = list(self.class_to_index.keys())
def __len__(self):
return len(self.class_to_index)
def __str__(self):
return f"<LabelEncoder(num_classes={len(self)})>"
def fit(self, y):
classes = np.unique(y)
for i, class_ in enumerate(classes):
self.class_to_index[class_] = i
self.index_to_class = {v: k for k, v in self.class_to_index.items()}
self.classes = list(self.class_to_index.keys())
return self
def encode(self, y):
encoded = np.zeros((len(y)), dtype=int)
for i, item in enumerate(y):
encoded[i] = self.class_to_index[item]
return encoded
def decode(self, y):
classes = []
for i, item in enumerate(y):
classes.append(self.index_to_class[item])
return classes
def save(self, fp):
with open(fp, 'w') as fp:
contents = {'class_to_index': self.class_to_index}
json.dump(contents, fp, indent=4, sort_keys=False)
@classmethod
def load(cls, fp):
with open(fp, 'r') as fp:
kwargs = json.load(fp=fp)
return cls(**kwargs)
# Encode
label_encoder = LabelEncoder()
label_encoder.fit(y_train)
NUM_CLASSES = len(label_encoder)
label_encoder.class_to_index
# Convert labels to tokens
print (f"y_train[0]: {y_train[0]}")
y_train = label_encoder.encode(y_train)
y_val = label_encoder.encode(y_val)
y_test = label_encoder.encode(y_test)
print (f"y_train[0]: {y_train[0]}")
# Class weights
counts = np.bincount(y_train)
class_weights = {i: 1.0/count for i, count in enumerate(counts)}
print (f"counts: {counts}\nweights: {class_weights}")
```
# Tokenizer
Our input data is text and we can't feed it directly to our models. So, we'll define a `Tokenizer` to convert our text input data into token indices. This means that every token (we can decide what a token is char, word, sub-word, etc.) is mapped to a unique index which allows us to represent our text as an array of indices.
```
import json
from collections import Counter
from more_itertools import take
class Tokenizer(object):
def __init__(self, char_level, num_tokens=None,
pad_token='<PAD>', oov_token='<UNK>',
token_to_index=None):
self.char_level = char_level
self.separator = '' if self.char_level else ' '
if num_tokens: num_tokens -= 2 # pad + unk tokens
self.num_tokens = num_tokens
self.pad_token = pad_token
self.oov_token = oov_token
if not token_to_index:
token_to_index = {pad_token: 0, oov_token: 1}
self.token_to_index = token_to_index
self.index_to_token = {v: k for k, v in self.token_to_index.items()}
def __len__(self):
return len(self.token_to_index)
def __str__(self):
return f"<Tokenizer(num_tokens={len(self)})>"
def fit_on_texts(self, texts):
if not self.char_level:
texts = [text.split(" ") for text in texts]
all_tokens = [token for text in texts for token in text]
counts = Counter(all_tokens).most_common(self.num_tokens)
self.min_token_freq = counts[-1][1]
for token, count in counts:
index = len(self)
self.token_to_index[token] = index
self.index_to_token[index] = token
return self
def texts_to_sequences(self, texts):
sequences = []
for text in texts:
if not self.char_level:
text = text.split(' ')
sequence = []
for token in text:
sequence.append(self.token_to_index.get(
token, self.token_to_index[self.oov_token]))
sequences.append(np.asarray(sequence))
return sequences
def sequences_to_texts(self, sequences):
texts = []
for sequence in sequences:
text = []
for index in sequence:
text.append(self.index_to_token.get(index, self.oov_token))
texts.append(self.separator.join([token for token in text]))
return texts
def save(self, fp):
with open(fp, 'w') as fp:
contents = {
'char_level': self.char_level,
'oov_token': self.oov_token,
'token_to_index': self.token_to_index
}
json.dump(contents, fp, indent=4, sort_keys=False)
@classmethod
def load(cls, fp):
with open(fp, 'r') as fp:
kwargs = json.load(fp=fp)
return cls(**kwargs)
```
We're going to restrict the number of tokens in our `Tokenizer` to the top 500 most frequent tokens (stop words already removed) because the full vocabulary size (~30K) is too large to run on Google Colab notebooks.
> It's important that we only fit using our train data split because during inference, our model will not always know every token so it's important to replicate that scenario with our validation and test splits as well.
```
# Tokenize
tokenizer = Tokenizer(char_level=False, num_tokens=500)
tokenizer.fit_on_texts(texts=X_train)
VOCAB_SIZE = len(tokenizer)
print (tokenizer)
# Sample of tokens
print (take(5, tokenizer.token_to_index.items()))
print (f"least freq token's freq: {tokenizer.min_token_freq}") # use this to adjust num_tokens
# Convert texts to sequences of indices
X_train = tokenizer.texts_to_sequences(X_train)
X_val = tokenizer.texts_to_sequences(X_val)
X_test = tokenizer.texts_to_sequences(X_test)
preprocessed_text = tokenizer.sequences_to_texts([X_train[0]])[0]
print ("Text to indices:\n"
f" (preprocessed) → {preprocessed_text}\n"
f" (tokenized) → {X_train[0]}")
```
# One-hot encoding
One-hot encoding creates a binary column for each unique value for the feature we're trying to map. All of the values in each token's array will be 0 except at the index that this specific token is represented by.
There are 5 words in the vocabulary:
```json
{
"a": 0,
"e": 1,
"i": 2,
"o": 3,
"u": 4
}
```
Then the text `aou` would be represented by:
```python
[[1. 0. 0. 0. 0.]
[0. 0. 0. 1. 0.]
[0. 0. 0. 0. 1.]]
```
One-hot encoding allows us to represent our data in a way that our models can process the data and isn't biased by the actual value of the token (ex. if your labels were actual numbers).
> We have already applied one-hot encoding in the previous lessons when we encoded our labels. Each label was represented by a unique index but when determining loss, we effectively use it's one hot representation and compared it to the predicted probability distribution. We never explicitly wrote this out since all of our previous tasks were multi-class which means every input had just one output class, so the 0s didn't affect the loss (though it did matter during back propagation).
```
def to_categorical(seq, num_classes):
"""One-hot encode a sequence of tokens."""
one_hot = np.zeros((len(seq), num_classes))
for i, item in enumerate(seq):
one_hot[i, item] = 1.
return one_hot
# One-hot encoding
print (X_train[0])
print (len(X_train[0]))
cat = to_categorical(seq=X_train[0], num_classes=len(tokenizer))
print (cat)
print (cat.shape)
# Convert tokens to one-hot
vocab_size = len(tokenizer)
X_train = [to_categorical(seq, num_classes=vocab_size) for seq in X_train]
X_val = [to_categorical(seq, num_classes=vocab_size) for seq in X_val]
X_test = [to_categorical(seq, num_classes=vocab_size) for seq in X_test]
```
# Padding
Our inputs are all of varying length but we need each batch to be uniformly shaped. Therefore, we will use padding to make all the inputs in the batch the same length. Our padding index will be 0 (note that this is consistent with the `<PAD>` token defined in our `Tokenizer`).
> One-hot encoding creates a batch of shape (`N`, `max_seq_len`, `vocab_size`) so we'll need to be able to pad 3D sequences.
```
def pad_sequences(sequences, max_seq_len=0):
"""Pad sequences to max length in sequence."""
max_seq_len = max(max_seq_len, max(len(sequence) for sequence in sequences))
num_classes = sequences[0].shape[-1]
padded_sequences = np.zeros((len(sequences), max_seq_len, num_classes))
for i, sequence in enumerate(sequences):
padded_sequences[i][:len(sequence)] = sequence
return padded_sequences
# 3D sequences
print (X_train[0].shape, X_train[1].shape, X_train[2].shape)
padded = pad_sequences(X_train[0:3])
print (padded.shape)
```
# Dataset
We're going to place our data into a [`Dataset`](https://pytorch.org/docs/stable/data.html#torch.utils.data.Dataset) and use a [`DataLoader`](https://pytorch.org/docs/stable/data.html#torch.utils.data.DataLoader) to efficiently create batches for training and evaluation.
```
FILTER_SIZE = 1 # unigram
class Dataset(torch.utils.data.Dataset):
def __init__(self, X, y, max_filter_size):
self.X = X
self.y = y
self.max_filter_size = max_filter_size
def __len__(self):
return len(self.y)
def __str__(self):
return f"<Dataset(N={len(self)})>"
def __getitem__(self, index):
X = self.X[index]
y = self.y[index]
return [X, y]
def collate_fn(self, batch):
"""Processing on a batch."""
# Get inputs
batch = np.array(batch, dtype=object)
X = batch[:, 0]
y = np.stack(batch[:, 1], axis=0)
# Pad sequences
X = pad_sequences(X, max_seq_len=self.max_filter_size)
# Cast
X = torch.FloatTensor(X.astype(np.int32))
y = torch.LongTensor(y.astype(np.int32))
return X, y
def create_dataloader(self, batch_size, shuffle=False, drop_last=False):
return torch.utils.data.DataLoader(
dataset=self, batch_size=batch_size, collate_fn=self.collate_fn,
shuffle=shuffle, drop_last=drop_last, pin_memory=True)
# Create datasets for embedding
train_dataset = Dataset(X=X_train, y=y_train, max_filter_size=FILTER_SIZE)
val_dataset = Dataset(X=X_val, y=y_val, max_filter_size=FILTER_SIZE)
test_dataset = Dataset(X=X_test, y=y_test, max_filter_size=FILTER_SIZE)
print ("Datasets:\n"
f" Train dataset:{train_dataset.__str__()}\n"
f" Val dataset: {val_dataset.__str__()}\n"
f" Test dataset: {test_dataset.__str__()}\n"
"Sample point:\n"
f" X: {test_dataset[0][0]}\n"
f" y: {test_dataset[0][1]}")
# Create dataloaders
batch_size = 64
train_dataloader = train_dataset.create_dataloader(batch_size=batch_size)
val_dataloader = val_dataset.create_dataloader(batch_size=batch_size)
test_dataloader = test_dataset.create_dataloader(batch_size=batch_size)
batch_X, batch_y = next(iter(test_dataloader))
print ("Sample batch:\n"
f" X: {list(batch_X.size())}\n"
f" y: {list(batch_y.size())}\n"
"Sample point:\n"
f" X: {batch_X[0]}\n"
f" y: {batch_y[0]}")
```
# CNN
## Inputs
We're going to learn about CNNs by applying them on 1D text data. In the dummy example below, our inputs are composed of character tokens that are one-hot encoded. We have a batch of N samples, where each sample has 8 characters and each character is represented by an array of 10 values (`vocab size=10`). This gives our inputs the size `(N, 8, 10)`.
> With PyTorch, when dealing with convolution, our inputs (X) need to have the channels as the second dimension, so our inputs will be `(N, 10, 8)`.
```
import math
import torch
import torch.nn as nn
import torch.nn.functional as F
# Assume all our inputs are padded to have the same # of words
batch_size = 64
max_seq_len = 8 # words per input
vocab_size = 10 # one hot size
x = torch.randn(batch_size, max_seq_len, vocab_size)
print(f"X: {x.shape}")
x = x.transpose(1, 2)
print(f"X: {x.shape}")
```
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/cnn/inputs.png" width="500">
</div>
This diagram above is for char-level tokens but extends to any level of tokenization (word-level in our case).
## Filters
At the core of CNNs are filters (aka weights, kernels, etc.) which convolve (slide) across our input to extract relevant features. The filters are initialized randomly but learn to pick up meaningful features from the input that aid in optimizing for the objective. The intuition here is that each filter represents a feature and we will use this filter on other inputs to capture the same feature (feature extraction via parameter sharing).
We can see convolution in the diagram below where we simplified the filters and inputs to be 2D for ease of visualization. Also note that the values are 0/1s but in reality they can be any floating point value.
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/cnn/convolution.gif" width="500">
</div>
Now let's return to our actual inputs `x`, which is of shape (8, 10) [`max_seq_len`, `vocab_size`] and we want to convolve on this input using filters. We will use 50 filters that are of size (1, 3) and has the same depth as the number of channels (`num_channels` = `vocab_size` = `one_hot_size` = 10). This gives our filter a shape of (3, 10, 50) [`kernel_size`, `vocab_size`, `num_filters`]
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/cnn/filters.png" width="500">
</div>
* **stride**: amount the filters move from one convolution operation to the next.
* **padding**: values (typically zero) padded to the input, typically to create a volume with whole number dimensions.
So far we've used a `stride` of 1 and `VALID` padding (no padding) but let's look at an example with a higher stride and difference between different padding approaches.
Padding types:
* **VALID**: no padding, the filters only use the "valid" values in the input. If the filter cannot reach all the input values (filters go left to right), the extra values on the right are dropped.
* **SAME**: adds padding evenly to the right (preferred) and left sides of the input so that all values in the input are processed.
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/cnn/padding.png" width="500">
</div>
We're going to use the [Conv1d](https://pytorch.org/docs/stable/generated/torch.nn.Conv1d.html#torch.nn.Conv1d) layer to process our inputs.
```
# Convolutional filters (VALID padding)
vocab_size = 10 # one hot size
num_filters = 50 # num filters
filter_size = 3 # filters are 3X3
stride = 1
padding = 0 # valid padding (no padding)
conv1 = nn.Conv1d(in_channels=vocab_size, out_channels=num_filters,
kernel_size=filter_size, stride=stride,
padding=padding, padding_mode='zeros')
print("conv: {}".format(conv1.weight.shape))
# Forward pass
z = conv1(x)
print (f"z: {z.shape}")
```
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/cnn/conv.png" width="700">
</div>
When we apply these filter on our inputs, we receive an output of shape (N, 6, 50). We get 50 for the output channel dim because we used 50 filters and 6 for the conv outputs because:
$W_1 = \frac{W_2 - F + 2P}{S} + 1 = \frac{8 - 3 + 2(0)}{1} + 1 = 6$
$H_1 = \frac{H_2 - F + 2P}{S} + 1 = \frac{1 - 1 + 2(0)}{1} + 1 = 1$
$D_2 = D_1 $
where:
* `W`: width of each input = 8
* `H`: height of each input = 1
* `D`: depth (# channels)
* `F`: filter size = 3
* `P`: padding = 0
* `S`: stride = 1
Now we'll add padding so that the convolutional outputs are the same shape as our inputs. The amount of padding for the `SAME` padding can be determined using the same equation. We want out output to have the same width as our input, so we solve for P:
$ \frac{W-F+2P}{S} + 1 = W $
$ P = \frac{S(W-1) - W + F}{2} $
If $P$ is not a whole number, we round up (using `math.ceil`) and place the extra padding on the right side.
```
# Convolutional filters (SAME padding)
vocab_size = 10 # one hot size
num_filters = 50 # num filters
filter_size = 3 # filters are 3X3
stride = 1
conv = nn.Conv1d(in_channels=vocab_size, out_channels=num_filters,
kernel_size=filter_size, stride=stride)
print("conv: {}".format(conv.weight.shape))
# `SAME` padding
padding_left = int((conv.stride[0]*(max_seq_len-1) - max_seq_len + filter_size)/2)
padding_right = int(math.ceil((conv.stride[0]*(max_seq_len-1) - max_seq_len + filter_size)/2))
print (f"padding: {(padding_left, padding_right)}")
# Forward pass
z = conv(F.pad(x, (padding_left, padding_right)))
print (f"z: {z.shape}")
```
> We will explore larger dimensional convolution layers in subsequent lessons. For example, [Conv2D](https://pytorch.org/docs/stable/generated/torch.nn.Conv2d.html#torch.nn.Conv2d) is used with 3D inputs (images, char-level text, etc.) and [Conv3D](https://pytorch.org/docs/stable/generated/torch.nn.Conv3d.html#torch.nn.Conv3d) is used for 4D inputs (videos, time-series, etc.).
## Pooling
The result of convolving filters on an input is a feature map. Due to the nature of convolution and overlaps, our feature map will have lots of redundant information. Pooling is a way to summarize a high-dimensional feature map into a lower dimensional one for simplified downstream computation. The pooling operation can be the max value, average, etc. in a certain receptive field. Below is an example of pooling where the outputs from a conv layer are `4X4` and we're going to apply max pool filters of size `2X2`.
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/cnn/pooling.png" width="500">
</div>
$W_2 = \frac{W_1 - F}{S} + 1 = \frac{4 - 2}{2} + 1 = 2$
$H_2 = \frac{H_1 - F}{S} + 1 = \frac{4 - 2}{2} + 1 = 2$
$ D_2 = D_1 $
where:
* `W`: width of each input = 4
* `H`: height of each input = 4
* `D`: depth (# channels)
* `F`: filter size = 2
* `S`: stride = 2
In our use case, we want to just take the one max value so we will use the [MaxPool1D](https://pytorch.org/docs/stable/generated/torch.nn.MaxPool1d.html#torch.nn.MaxPool1d) layer, so our max-pool filter size will be max_seq_len.
```
# Max pooling
pool_output = F.max_pool1d(z, z.size(2))
print("Size: {}".format(pool_output.shape))
```
## Batch Normalization
The last topic we'll cover before constructing our model is [batch normalization](https://arxiv.org/abs/1502.03167). It's an operation that will standardize (mean=0, std=1) the activations from the previous layer. Recall that we used to standardize our inputs in previous notebooks so our model can optimize quickly with larger learning rates. It's the same concept here but we continue to maintain standardized values throughout the forward pass to further aid optimization.
```
# Batch normalization
batch_norm = nn.BatchNorm1d(num_features=num_filters)
z = batch_norm(conv(x)) # applied to activations (after conv layer & before pooling)
print (f"z: {z.shape}")
# Mean and std before batchnorm
print (f"mean: {torch.mean(conv1(x)):.2f}, std: {torch.std(conv(x)):.2f}")
# Mean and std after batchnorm
print (f"mean: {torch.mean(z):.2f}, std: {torch.std(z):.2f}")
```
# Modeling
## Model
Let's visualize the model's forward pass.
1. We'll first tokenize our inputs (`batch_size`, `max_seq_len`).
2. Then we'll one-hot encode our tokenized inputs (`batch_size`, `max_seq_len`, `vocab_size`).
3. We'll apply convolution via filters (`filter_size`, `vocab_size`, `num_filters`) followed by batch normalization. Our filters act as character level n-gram detectors.
4. We'll apply 1D global max pooling which will extract the most relevant information from the feature maps for making the decision.
5. We feed the pool outputs to a fully-connected (FC) layer (with dropout).
6. We use one more FC layer with softmax to derive class probabilities.
<div align="left">
<img src="https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/images/basics/cnn/model.png" width="1000">
</div>
```
NUM_FILTERS = 50
HIDDEN_DIM = 100
DROPOUT_P = 0.1
class CNN(nn.Module):
def __init__(self, vocab_size, num_filters, filter_size,
hidden_dim, dropout_p, num_classes):
super(CNN, self).__init__()
# Convolutional filters
self.filter_size = filter_size
self.conv = nn.Conv1d(
in_channels=vocab_size, out_channels=num_filters,
kernel_size=filter_size, stride=1, padding=0, padding_mode='zeros')
self.batch_norm = nn.BatchNorm1d(num_features=num_filters)
# FC layers
self.fc1 = nn.Linear(num_filters, hidden_dim)
self.dropout = nn.Dropout(dropout_p)
self.fc2 = nn.Linear(hidden_dim, num_classes)
def forward(self, inputs, channel_first=False, apply_softmax=False):
# Rearrange input so num_channels is in dim 1 (N, C, L)
x_in, = inputs
if not channel_first:
x_in = x_in.transpose(1, 2)
# Padding for `SAME` padding
max_seq_len = x_in.shape[2]
padding_left = int((self.conv.stride[0]*(max_seq_len-1) - max_seq_len + self.filter_size)/2)
padding_right = int(math.ceil((self.conv.stride[0]*(max_seq_len-1) - max_seq_len + self.filter_size)/2))
# Conv outputs
z = self.conv(F.pad(x_in, (padding_left, padding_right)))
z = F.max_pool1d(z, z.size(2)).squeeze(2)
# FC layer
z = self.fc1(z)
z = self.dropout(z)
y_pred = self.fc2(z)
if apply_softmax:
y_pred = F.softmax(y_pred, dim=1)
return y_pred
# Initialize model
model = CNN(vocab_size=VOCAB_SIZE, num_filters=NUM_FILTERS, filter_size=FILTER_SIZE,
hidden_dim=HIDDEN_DIM, dropout_p=DROPOUT_P, num_classes=NUM_CLASSES)
model = model.to(device) # set device
print (model.named_parameters)
```
> We used `SAME` padding (w/ stride=1) which means that the conv outputs will have the same width (`max_seq_len`) as our inputs. The amount of padding differs for each batch based on the `max_seq_len` but you can calculate it by solving for P in the equation below.
$ \frac{W_1 - F + 2P}{S} + 1 = W_2 $
$ \frac{\text{max_seq_len } - \text{ filter_size } + 2P}{\text{stride}} + 1 = \text{max_seq_len} $
$ P = \frac{\text{stride}(\text{max_seq_len}-1) - \text{max_seq_len} + \text{filter_size}}{2} $
If $P$ is not a whole number, we round up (using `math.ceil`) and place the extra padding on the right side.
## Training
Let's create the `Trainer` class that we'll use to facilitate training for our experiments. Notice that we're now moving the `train` function inside this class.
```
from torch.optim import Adam
LEARNING_RATE = 1e-3
PATIENCE = 5
NUM_EPOCHS = 10
class Trainer(object):
def __init__(self, model, device, loss_fn=None, optimizer=None, scheduler=None):
# Set params
self.model = model
self.device = device
self.loss_fn = loss_fn
self.optimizer = optimizer
self.scheduler = scheduler
def train_step(self, dataloader):
"""Train step."""
# Set model to train mode
self.model.train()
loss = 0.0
# Iterate over train batches
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, targets = batch[:-1], batch[-1]
self.optimizer.zero_grad() # Reset gradients
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, targets) # Define loss
J.backward() # Backward pass
self.optimizer.step() # Update weights
# Cumulative Metrics
loss += (J.detach().item() - loss) / (i + 1)
return loss
def eval_step(self, dataloader):
"""Validation or test step."""
# Set model to eval mode
self.model.eval()
loss = 0.0
y_trues, y_probs = [], []
# Iterate over val batches
with torch.no_grad():
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, y_true = batch[:-1], batch[-1]
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, y_true).item()
# Cumulative Metrics
loss += (J - loss) / (i + 1)
# Store outputs
y_prob = torch.sigmoid(z).cpu().numpy()
y_probs.extend(y_prob)
y_trues.extend(y_true.cpu().numpy())
return loss, np.vstack(y_trues), np.vstack(y_probs)
def predict_step(self, dataloader):
"""Prediction step."""
# Set model to eval mode
self.model.eval()
y_probs = []
# Iterate over val batches
with torch.no_grad():
for i, batch in enumerate(dataloader):
# Forward pass w/ inputs
inputs, targets = batch[:-1], batch[-1]
y_prob = self.model(inputs, apply_softmax=True)
# Store outputs
y_probs.extend(y_prob)
return np.vstack(y_probs)
def train(self, num_epochs, patience, train_dataloader, val_dataloader):
best_val_loss = np.inf
for epoch in range(num_epochs):
# Steps
train_loss = self.train_step(dataloader=train_dataloader)
val_loss, _, _ = self.eval_step(dataloader=val_dataloader)
self.scheduler.step(val_loss)
# Early stopping
if val_loss < best_val_loss:
best_val_loss = val_loss
best_model = self.model
_patience = patience # reset _patience
else:
_patience -= 1
if not _patience: # 0
print("Stopping early!")
break
# Logging
print(
f"Epoch: {epoch+1} | "
f"train_loss: {train_loss:.5f}, "
f"val_loss: {val_loss:.5f}, "
f"lr: {self.optimizer.param_groups[0]['lr']:.2E}, "
f"_patience: {_patience}"
)
return best_model
# Define Loss
class_weights_tensor = torch.Tensor(list(class_weights.values())).to(device)
loss = nn.CrossEntropyLoss(weight=class_weights_tensor)
# Define optimizer & scheduler
optimizer = Adam(model.parameters(), lr=LEARNING_RATE)
scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(
optimizer, mode='min', factor=0.1, patience=3)
# Trainer module
trainer = Trainer(
model=model, device=device, loss_fn=loss_fn,
optimizer=optimizer, scheduler=scheduler)
# Train
best_model = trainer.train(
NUM_EPOCHS, PATIENCE, train_dataloader, val_dataloader)
```
## Evaluation
```
import json
from pathlib import Path
from sklearn.metrics import precision_recall_fscore_support
def get_performance(y_true, y_pred, classes):
"""Per-class performance metrics."""
# Performance
performance = {"overall": {}, "class": {}}
# Overall performance
metrics = precision_recall_fscore_support(y_true, y_pred, average="weighted")
performance["overall"]["precision"] = metrics[0]
performance["overall"]["recall"] = metrics[1]
performance["overall"]["f1"] = metrics[2]
performance["overall"]["num_samples"] = np.float64(len(y_true))
# Per-class performance
metrics = precision_recall_fscore_support(y_true, y_pred, average=None)
for i in range(len(classes)):
performance["class"][classes[i]] = {
"precision": metrics[0][i],
"recall": metrics[1][i],
"f1": metrics[2][i],
"num_samples": np.float64(metrics[3][i]),
}
return performance
# Get predictions
test_loss, y_true, y_prob = trainer.eval_step(dataloader=test_dataloader)
y_pred = np.argmax(y_prob, axis=1)
# Determine performance
performance = get_performance(
y_true=y_test, y_pred=y_pred, classes=label_encoder.classes)
print (json.dumps(performance['overall'], indent=2))
# Save artifacts
dir = Path("cnn")
dir.mkdir(parents=True, exist_ok=True)
label_encoder.save(fp=Path(dir, 'label_encoder.json'))
tokenizer.save(fp=Path(dir, 'tokenizer.json'))
torch.save(best_model.state_dict(), Path(dir, 'model.pt'))
with open(Path(dir, 'performance.json'), "w") as fp:
json.dump(performance, indent=2, sort_keys=False, fp=fp)
```
## Inference
```
def get_probability_distribution(y_prob, classes):
"""Create a dict of class probabilities from an array."""
results = {}
for i, class_ in enumerate(classes):
results[class_] = np.float64(y_prob[i])
sorted_results = {k: v for k, v in sorted(
results.items(), key=lambda item: item[1], reverse=True)}
return sorted_results
# Load artifacts
device = torch.device("cpu")
label_encoder = LabelEncoder.load(fp=Path(dir, 'label_encoder.json'))
tokenizer = Tokenizer.load(fp=Path(dir, 'tokenizer.json'))
model = CNN(
vocab_size=VOCAB_SIZE, num_filters=NUM_FILTERS, filter_size=FILTER_SIZE,
hidden_dim=HIDDEN_DIM, dropout_p=DROPOUT_P, num_classes=NUM_CLASSES)
model.load_state_dict(torch.load(Path(dir, 'model.pt'), map_location=device))
model.to(device)
# Initialize trainer
trainer = Trainer(model=model, device=device)
# Dataloader
text = "What a day for the new york stock market to go bust!"
sequences = tokenizer.texts_to_sequences([preprocess(text)])
print (tokenizer.sequences_to_texts(sequences))
X = [to_categorical(seq, num_classes=len(tokenizer)) for seq in sequences]
y_filler = label_encoder.encode([label_encoder.classes[0]]*len(X))
dataset = Dataset(X=X, y=y_filler, max_filter_size=FILTER_SIZE)
dataloader = dataset.create_dataloader(batch_size=batch_size)
# Inference
y_prob = trainer.predict_step(dataloader)
y_pred = np.argmax(y_prob, axis=1)
label_encoder.decode(y_pred)
# Class distributions
prob_dist = get_probability_distribution(y_prob=y_prob[0], classes=label_encoder.classes)
print (json.dumps(prob_dist, indent=2))
```
# Interpretability
We went through all the trouble of padding our inputs before convolution to result is outputs of the same shape as our inputs so we can try to get some interpretability. Since every token is mapped to a convolutional output on whcih we apply max pooling, we can see which token's output was most influential towards the prediction. We first need to get the conv outputs from our model:
```
import collections
import seaborn as sns
class InterpretableCNN(nn.Module):
def __init__(self, vocab_size, num_filters, filter_size,
hidden_dim, dropout_p, num_classes):
super(InterpretableCNN, self).__init__()
# Convolutional filters
self.filter_size = filter_size
self.conv = nn.Conv1d(
in_channels=vocab_size, out_channels=num_filters,
kernel_size=filter_size, stride=1, padding=0, padding_mode='zeros')
self.batch_norm = nn.BatchNorm1d(num_features=num_filters)
# FC layers
self.fc1 = nn.Linear(num_filters, hidden_dim)
self.dropout = nn.Dropout(dropout_p)
self.fc2 = nn.Linear(hidden_dim, num_classes)
def forward(self, inputs, channel_first=False, apply_softmax=False):
# Rearrange input so num_channels is in dim 1 (N, C, L)
x_in, = inputs
if not channel_first:
x_in = x_in.transpose(1, 2)
# Padding for `SAME` padding
max_seq_len = x_in.shape[2]
padding_left = int((self.conv.stride[0]*(max_seq_len-1) - max_seq_len + self.filter_size)/2)
padding_right = int(math.ceil((self.conv.stride[0]*(max_seq_len-1) - max_seq_len + self.filter_size)/2))
# Conv outputs
z = self.conv(F.pad(x_in, (padding_left, padding_right)))
return z
# Initialize
interpretable_model = InterpretableCNN(
vocab_size=len(tokenizer), num_filters=NUM_FILTERS, filter_size=FILTER_SIZE,
hidden_dim=HIDDEN_DIM, dropout_p=DROPOUT_P, num_classes=NUM_CLASSES)
# Load weights (same architecture)
interpretable_model.load_state_dict(torch.load(Path(dir, 'model.pt'), map_location=device))
interpretable_model.to(device)
# Initialize trainer
interpretable_trainer = Trainer(model=interpretable_model, device=device)
# Get conv outputs
conv_outputs = interpretable_trainer.predict_step(dataloader)
print (conv_outputs.shape) # (num_filters, max_seq_len)
# Visualize a bi-gram filter's outputs
tokens = tokenizer.sequences_to_texts(sequences)[0].split(' ')
sns.heatmap(conv_outputs, xticklabels=tokens)
```
The filters have high values for the words `stock` and `market` which influenced the `Business` category classification.
> This is a crude technique (maxpool doesn't strictly behave this way on a batch) loosely based off of more elaborate [interpretability](https://arxiv.org/abs/1312.6034) methods.
|
github_jupyter
|
import numpy as np
import pandas as pd
import random
import torch
import torch.nn as nn
SEED = 1234
def set_seeds(seed=1234):
"""Set seeds for reproducibility."""
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed) # multi-GPU
# Set seeds for reproducibility
set_seeds(seed=SEED)
# Set device
cuda = True
device = torch.device('cuda' if (
torch.cuda.is_available() and cuda) else 'cpu')
torch.set_default_tensor_type('torch.FloatTensor')
if device.type == 'cuda':
torch.set_default_tensor_type('torch.cuda.FloatTensor')
print (device)
# Load data
url = "https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/datasets/news.csv"
df = pd.read_csv(url, header=0) # load
df = df.sample(frac=1).reset_index(drop=True) # shuffle
df.head()
import nltk
from nltk.corpus import stopwords
from nltk.stem import PorterStemmer
import re
nltk.download('stopwords')
STOPWORDS = stopwords.words('english')
print (STOPWORDS[:5])
porter = PorterStemmer()
def preprocess(text, stopwords=STOPWORDS):
"""Conditional preprocessing on our text unique to our task."""
# Lower
text = text.lower()
# Remove stopwords
pattern = re.compile(r'\b(' + r'|'.join(stopwords) + r')\b\s*')
text = pattern.sub('', text)
# Remove words in paranthesis
text = re.sub(r'\([^)]*\)', '', text)
# Spacing and filters
text = re.sub(r"([-;;.,!?<=>])", r" \1 ", text)
text = re.sub('[^A-Za-z0-9]+', ' ', text) # remove non alphanumeric chars
text = re.sub(' +', ' ', text) # remove multiple spaces
text = text.strip()
return text
# Sample
text = "Great week for the NYSE!"
preprocess(text=text)
# Apply to dataframe
preprocessed_df = df.copy()
preprocessed_df.title = preprocessed_df.title.apply(preprocess)
print (f"{df.title.values[0]}\n\n{preprocessed_df.title.values[0]}")
import collections
from sklearn.model_selection import train_test_split
TRAIN_SIZE = 0.7
VAL_SIZE = 0.15
TEST_SIZE = 0.15
def train_val_test_split(X, y, train_size):
"""Split dataset into data splits."""
X_train, X_, y_train, y_ = train_test_split(X, y, train_size=TRAIN_SIZE, stratify=y)
X_val, X_test, y_val, y_test = train_test_split(X_, y_, train_size=0.5, stratify=y_)
return X_train, X_val, X_test, y_train, y_val, y_test
# Data
X = preprocessed_df["title"].values
y = preprocessed_df["category"].values
# Create data splits
X_train, X_val, X_test, y_train, y_val, y_test = train_val_test_split(
X=X, y=y, train_size=TRAIN_SIZE)
print (f"X_train: {X_train.shape}, y_train: {y_train.shape}")
print (f"X_val: {X_val.shape}, y_val: {y_val.shape}")
print (f"X_test: {X_test.shape}, y_test: {y_test.shape}")
print (f"Sample point: {X_train[0]} → {y_train[0]}")
import itertools
class LabelEncoder(object):
"""Label encoder for tag labels."""
def __init__(self, class_to_index={}):
self.class_to_index = class_to_index
self.index_to_class = {v: k for k, v in self.class_to_index.items()}
self.classes = list(self.class_to_index.keys())
def __len__(self):
return len(self.class_to_index)
def __str__(self):
return f"<LabelEncoder(num_classes={len(self)})>"
def fit(self, y):
classes = np.unique(y)
for i, class_ in enumerate(classes):
self.class_to_index[class_] = i
self.index_to_class = {v: k for k, v in self.class_to_index.items()}
self.classes = list(self.class_to_index.keys())
return self
def encode(self, y):
encoded = np.zeros((len(y)), dtype=int)
for i, item in enumerate(y):
encoded[i] = self.class_to_index[item]
return encoded
def decode(self, y):
classes = []
for i, item in enumerate(y):
classes.append(self.index_to_class[item])
return classes
def save(self, fp):
with open(fp, 'w') as fp:
contents = {'class_to_index': self.class_to_index}
json.dump(contents, fp, indent=4, sort_keys=False)
@classmethod
def load(cls, fp):
with open(fp, 'r') as fp:
kwargs = json.load(fp=fp)
return cls(**kwargs)
# Encode
label_encoder = LabelEncoder()
label_encoder.fit(y_train)
NUM_CLASSES = len(label_encoder)
label_encoder.class_to_index
# Convert labels to tokens
print (f"y_train[0]: {y_train[0]}")
y_train = label_encoder.encode(y_train)
y_val = label_encoder.encode(y_val)
y_test = label_encoder.encode(y_test)
print (f"y_train[0]: {y_train[0]}")
# Class weights
counts = np.bincount(y_train)
class_weights = {i: 1.0/count for i, count in enumerate(counts)}
print (f"counts: {counts}\nweights: {class_weights}")
import json
from collections import Counter
from more_itertools import take
class Tokenizer(object):
def __init__(self, char_level, num_tokens=None,
pad_token='<PAD>', oov_token='<UNK>',
token_to_index=None):
self.char_level = char_level
self.separator = '' if self.char_level else ' '
if num_tokens: num_tokens -= 2 # pad + unk tokens
self.num_tokens = num_tokens
self.pad_token = pad_token
self.oov_token = oov_token
if not token_to_index:
token_to_index = {pad_token: 0, oov_token: 1}
self.token_to_index = token_to_index
self.index_to_token = {v: k for k, v in self.token_to_index.items()}
def __len__(self):
return len(self.token_to_index)
def __str__(self):
return f"<Tokenizer(num_tokens={len(self)})>"
def fit_on_texts(self, texts):
if not self.char_level:
texts = [text.split(" ") for text in texts]
all_tokens = [token for text in texts for token in text]
counts = Counter(all_tokens).most_common(self.num_tokens)
self.min_token_freq = counts[-1][1]
for token, count in counts:
index = len(self)
self.token_to_index[token] = index
self.index_to_token[index] = token
return self
def texts_to_sequences(self, texts):
sequences = []
for text in texts:
if not self.char_level:
text = text.split(' ')
sequence = []
for token in text:
sequence.append(self.token_to_index.get(
token, self.token_to_index[self.oov_token]))
sequences.append(np.asarray(sequence))
return sequences
def sequences_to_texts(self, sequences):
texts = []
for sequence in sequences:
text = []
for index in sequence:
text.append(self.index_to_token.get(index, self.oov_token))
texts.append(self.separator.join([token for token in text]))
return texts
def save(self, fp):
with open(fp, 'w') as fp:
contents = {
'char_level': self.char_level,
'oov_token': self.oov_token,
'token_to_index': self.token_to_index
}
json.dump(contents, fp, indent=4, sort_keys=False)
@classmethod
def load(cls, fp):
with open(fp, 'r') as fp:
kwargs = json.load(fp=fp)
return cls(**kwargs)
# Tokenize
tokenizer = Tokenizer(char_level=False, num_tokens=500)
tokenizer.fit_on_texts(texts=X_train)
VOCAB_SIZE = len(tokenizer)
print (tokenizer)
# Sample of tokens
print (take(5, tokenizer.token_to_index.items()))
print (f"least freq token's freq: {tokenizer.min_token_freq}") # use this to adjust num_tokens
# Convert texts to sequences of indices
X_train = tokenizer.texts_to_sequences(X_train)
X_val = tokenizer.texts_to_sequences(X_val)
X_test = tokenizer.texts_to_sequences(X_test)
preprocessed_text = tokenizer.sequences_to_texts([X_train[0]])[0]
print ("Text to indices:\n"
f" (preprocessed) → {preprocessed_text}\n"
f" (tokenized) → {X_train[0]}")
{
"a": 0,
"e": 1,
"i": 2,
"o": 3,
"u": 4
}
[[1. 0. 0. 0. 0.]
[0. 0. 0. 1. 0.]
[0. 0. 0. 0. 1.]]
def to_categorical(seq, num_classes):
"""One-hot encode a sequence of tokens."""
one_hot = np.zeros((len(seq), num_classes))
for i, item in enumerate(seq):
one_hot[i, item] = 1.
return one_hot
# One-hot encoding
print (X_train[0])
print (len(X_train[0]))
cat = to_categorical(seq=X_train[0], num_classes=len(tokenizer))
print (cat)
print (cat.shape)
# Convert tokens to one-hot
vocab_size = len(tokenizer)
X_train = [to_categorical(seq, num_classes=vocab_size) for seq in X_train]
X_val = [to_categorical(seq, num_classes=vocab_size) for seq in X_val]
X_test = [to_categorical(seq, num_classes=vocab_size) for seq in X_test]
def pad_sequences(sequences, max_seq_len=0):
"""Pad sequences to max length in sequence."""
max_seq_len = max(max_seq_len, max(len(sequence) for sequence in sequences))
num_classes = sequences[0].shape[-1]
padded_sequences = np.zeros((len(sequences), max_seq_len, num_classes))
for i, sequence in enumerate(sequences):
padded_sequences[i][:len(sequence)] = sequence
return padded_sequences
# 3D sequences
print (X_train[0].shape, X_train[1].shape, X_train[2].shape)
padded = pad_sequences(X_train[0:3])
print (padded.shape)
FILTER_SIZE = 1 # unigram
class Dataset(torch.utils.data.Dataset):
def __init__(self, X, y, max_filter_size):
self.X = X
self.y = y
self.max_filter_size = max_filter_size
def __len__(self):
return len(self.y)
def __str__(self):
return f"<Dataset(N={len(self)})>"
def __getitem__(self, index):
X = self.X[index]
y = self.y[index]
return [X, y]
def collate_fn(self, batch):
"""Processing on a batch."""
# Get inputs
batch = np.array(batch, dtype=object)
X = batch[:, 0]
y = np.stack(batch[:, 1], axis=0)
# Pad sequences
X = pad_sequences(X, max_seq_len=self.max_filter_size)
# Cast
X = torch.FloatTensor(X.astype(np.int32))
y = torch.LongTensor(y.astype(np.int32))
return X, y
def create_dataloader(self, batch_size, shuffle=False, drop_last=False):
return torch.utils.data.DataLoader(
dataset=self, batch_size=batch_size, collate_fn=self.collate_fn,
shuffle=shuffle, drop_last=drop_last, pin_memory=True)
# Create datasets for embedding
train_dataset = Dataset(X=X_train, y=y_train, max_filter_size=FILTER_SIZE)
val_dataset = Dataset(X=X_val, y=y_val, max_filter_size=FILTER_SIZE)
test_dataset = Dataset(X=X_test, y=y_test, max_filter_size=FILTER_SIZE)
print ("Datasets:\n"
f" Train dataset:{train_dataset.__str__()}\n"
f" Val dataset: {val_dataset.__str__()}\n"
f" Test dataset: {test_dataset.__str__()}\n"
"Sample point:\n"
f" X: {test_dataset[0][0]}\n"
f" y: {test_dataset[0][1]}")
# Create dataloaders
batch_size = 64
train_dataloader = train_dataset.create_dataloader(batch_size=batch_size)
val_dataloader = val_dataset.create_dataloader(batch_size=batch_size)
test_dataloader = test_dataset.create_dataloader(batch_size=batch_size)
batch_X, batch_y = next(iter(test_dataloader))
print ("Sample batch:\n"
f" X: {list(batch_X.size())}\n"
f" y: {list(batch_y.size())}\n"
"Sample point:\n"
f" X: {batch_X[0]}\n"
f" y: {batch_y[0]}")
import math
import torch
import torch.nn as nn
import torch.nn.functional as F
# Assume all our inputs are padded to have the same # of words
batch_size = 64
max_seq_len = 8 # words per input
vocab_size = 10 # one hot size
x = torch.randn(batch_size, max_seq_len, vocab_size)
print(f"X: {x.shape}")
x = x.transpose(1, 2)
print(f"X: {x.shape}")
# Convolutional filters (VALID padding)
vocab_size = 10 # one hot size
num_filters = 50 # num filters
filter_size = 3 # filters are 3X3
stride = 1
padding = 0 # valid padding (no padding)
conv1 = nn.Conv1d(in_channels=vocab_size, out_channels=num_filters,
kernel_size=filter_size, stride=stride,
padding=padding, padding_mode='zeros')
print("conv: {}".format(conv1.weight.shape))
# Forward pass
z = conv1(x)
print (f"z: {z.shape}")
# Convolutional filters (SAME padding)
vocab_size = 10 # one hot size
num_filters = 50 # num filters
filter_size = 3 # filters are 3X3
stride = 1
conv = nn.Conv1d(in_channels=vocab_size, out_channels=num_filters,
kernel_size=filter_size, stride=stride)
print("conv: {}".format(conv.weight.shape))
# `SAME` padding
padding_left = int((conv.stride[0]*(max_seq_len-1) - max_seq_len + filter_size)/2)
padding_right = int(math.ceil((conv.stride[0]*(max_seq_len-1) - max_seq_len + filter_size)/2))
print (f"padding: {(padding_left, padding_right)}")
# Forward pass
z = conv(F.pad(x, (padding_left, padding_right)))
print (f"z: {z.shape}")
# Max pooling
pool_output = F.max_pool1d(z, z.size(2))
print("Size: {}".format(pool_output.shape))
# Batch normalization
batch_norm = nn.BatchNorm1d(num_features=num_filters)
z = batch_norm(conv(x)) # applied to activations (after conv layer & before pooling)
print (f"z: {z.shape}")
# Mean and std before batchnorm
print (f"mean: {torch.mean(conv1(x)):.2f}, std: {torch.std(conv(x)):.2f}")
# Mean and std after batchnorm
print (f"mean: {torch.mean(z):.2f}, std: {torch.std(z):.2f}")
NUM_FILTERS = 50
HIDDEN_DIM = 100
DROPOUT_P = 0.1
class CNN(nn.Module):
def __init__(self, vocab_size, num_filters, filter_size,
hidden_dim, dropout_p, num_classes):
super(CNN, self).__init__()
# Convolutional filters
self.filter_size = filter_size
self.conv = nn.Conv1d(
in_channels=vocab_size, out_channels=num_filters,
kernel_size=filter_size, stride=1, padding=0, padding_mode='zeros')
self.batch_norm = nn.BatchNorm1d(num_features=num_filters)
# FC layers
self.fc1 = nn.Linear(num_filters, hidden_dim)
self.dropout = nn.Dropout(dropout_p)
self.fc2 = nn.Linear(hidden_dim, num_classes)
def forward(self, inputs, channel_first=False, apply_softmax=False):
# Rearrange input so num_channels is in dim 1 (N, C, L)
x_in, = inputs
if not channel_first:
x_in = x_in.transpose(1, 2)
# Padding for `SAME` padding
max_seq_len = x_in.shape[2]
padding_left = int((self.conv.stride[0]*(max_seq_len-1) - max_seq_len + self.filter_size)/2)
padding_right = int(math.ceil((self.conv.stride[0]*(max_seq_len-1) - max_seq_len + self.filter_size)/2))
# Conv outputs
z = self.conv(F.pad(x_in, (padding_left, padding_right)))
z = F.max_pool1d(z, z.size(2)).squeeze(2)
# FC layer
z = self.fc1(z)
z = self.dropout(z)
y_pred = self.fc2(z)
if apply_softmax:
y_pred = F.softmax(y_pred, dim=1)
return y_pred
# Initialize model
model = CNN(vocab_size=VOCAB_SIZE, num_filters=NUM_FILTERS, filter_size=FILTER_SIZE,
hidden_dim=HIDDEN_DIM, dropout_p=DROPOUT_P, num_classes=NUM_CLASSES)
model = model.to(device) # set device
print (model.named_parameters)
from torch.optim import Adam
LEARNING_RATE = 1e-3
PATIENCE = 5
NUM_EPOCHS = 10
class Trainer(object):
def __init__(self, model, device, loss_fn=None, optimizer=None, scheduler=None):
# Set params
self.model = model
self.device = device
self.loss_fn = loss_fn
self.optimizer = optimizer
self.scheduler = scheduler
def train_step(self, dataloader):
"""Train step."""
# Set model to train mode
self.model.train()
loss = 0.0
# Iterate over train batches
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, targets = batch[:-1], batch[-1]
self.optimizer.zero_grad() # Reset gradients
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, targets) # Define loss
J.backward() # Backward pass
self.optimizer.step() # Update weights
# Cumulative Metrics
loss += (J.detach().item() - loss) / (i + 1)
return loss
def eval_step(self, dataloader):
"""Validation or test step."""
# Set model to eval mode
self.model.eval()
loss = 0.0
y_trues, y_probs = [], []
# Iterate over val batches
with torch.no_grad():
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, y_true = batch[:-1], batch[-1]
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, y_true).item()
# Cumulative Metrics
loss += (J - loss) / (i + 1)
# Store outputs
y_prob = torch.sigmoid(z).cpu().numpy()
y_probs.extend(y_prob)
y_trues.extend(y_true.cpu().numpy())
return loss, np.vstack(y_trues), np.vstack(y_probs)
def predict_step(self, dataloader):
"""Prediction step."""
# Set model to eval mode
self.model.eval()
y_probs = []
# Iterate over val batches
with torch.no_grad():
for i, batch in enumerate(dataloader):
# Forward pass w/ inputs
inputs, targets = batch[:-1], batch[-1]
y_prob = self.model(inputs, apply_softmax=True)
# Store outputs
y_probs.extend(y_prob)
return np.vstack(y_probs)
def train(self, num_epochs, patience, train_dataloader, val_dataloader):
best_val_loss = np.inf
for epoch in range(num_epochs):
# Steps
train_loss = self.train_step(dataloader=train_dataloader)
val_loss, _, _ = self.eval_step(dataloader=val_dataloader)
self.scheduler.step(val_loss)
# Early stopping
if val_loss < best_val_loss:
best_val_loss = val_loss
best_model = self.model
_patience = patience # reset _patience
else:
_patience -= 1
if not _patience: # 0
print("Stopping early!")
break
# Logging
print(
f"Epoch: {epoch+1} | "
f"train_loss: {train_loss:.5f}, "
f"val_loss: {val_loss:.5f}, "
f"lr: {self.optimizer.param_groups[0]['lr']:.2E}, "
f"_patience: {_patience}"
)
return best_model
# Define Loss
class_weights_tensor = torch.Tensor(list(class_weights.values())).to(device)
loss = nn.CrossEntropyLoss(weight=class_weights_tensor)
# Define optimizer & scheduler
optimizer = Adam(model.parameters(), lr=LEARNING_RATE)
scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(
optimizer, mode='min', factor=0.1, patience=3)
# Trainer module
trainer = Trainer(
model=model, device=device, loss_fn=loss_fn,
optimizer=optimizer, scheduler=scheduler)
# Train
best_model = trainer.train(
NUM_EPOCHS, PATIENCE, train_dataloader, val_dataloader)
import json
from pathlib import Path
from sklearn.metrics import precision_recall_fscore_support
def get_performance(y_true, y_pred, classes):
"""Per-class performance metrics."""
# Performance
performance = {"overall": {}, "class": {}}
# Overall performance
metrics = precision_recall_fscore_support(y_true, y_pred, average="weighted")
performance["overall"]["precision"] = metrics[0]
performance["overall"]["recall"] = metrics[1]
performance["overall"]["f1"] = metrics[2]
performance["overall"]["num_samples"] = np.float64(len(y_true))
# Per-class performance
metrics = precision_recall_fscore_support(y_true, y_pred, average=None)
for i in range(len(classes)):
performance["class"][classes[i]] = {
"precision": metrics[0][i],
"recall": metrics[1][i],
"f1": metrics[2][i],
"num_samples": np.float64(metrics[3][i]),
}
return performance
# Get predictions
test_loss, y_true, y_prob = trainer.eval_step(dataloader=test_dataloader)
y_pred = np.argmax(y_prob, axis=1)
# Determine performance
performance = get_performance(
y_true=y_test, y_pred=y_pred, classes=label_encoder.classes)
print (json.dumps(performance['overall'], indent=2))
# Save artifacts
dir = Path("cnn")
dir.mkdir(parents=True, exist_ok=True)
label_encoder.save(fp=Path(dir, 'label_encoder.json'))
tokenizer.save(fp=Path(dir, 'tokenizer.json'))
torch.save(best_model.state_dict(), Path(dir, 'model.pt'))
with open(Path(dir, 'performance.json'), "w") as fp:
json.dump(performance, indent=2, sort_keys=False, fp=fp)
def get_probability_distribution(y_prob, classes):
"""Create a dict of class probabilities from an array."""
results = {}
for i, class_ in enumerate(classes):
results[class_] = np.float64(y_prob[i])
sorted_results = {k: v for k, v in sorted(
results.items(), key=lambda item: item[1], reverse=True)}
return sorted_results
# Load artifacts
device = torch.device("cpu")
label_encoder = LabelEncoder.load(fp=Path(dir, 'label_encoder.json'))
tokenizer = Tokenizer.load(fp=Path(dir, 'tokenizer.json'))
model = CNN(
vocab_size=VOCAB_SIZE, num_filters=NUM_FILTERS, filter_size=FILTER_SIZE,
hidden_dim=HIDDEN_DIM, dropout_p=DROPOUT_P, num_classes=NUM_CLASSES)
model.load_state_dict(torch.load(Path(dir, 'model.pt'), map_location=device))
model.to(device)
# Initialize trainer
trainer = Trainer(model=model, device=device)
# Dataloader
text = "What a day for the new york stock market to go bust!"
sequences = tokenizer.texts_to_sequences([preprocess(text)])
print (tokenizer.sequences_to_texts(sequences))
X = [to_categorical(seq, num_classes=len(tokenizer)) for seq in sequences]
y_filler = label_encoder.encode([label_encoder.classes[0]]*len(X))
dataset = Dataset(X=X, y=y_filler, max_filter_size=FILTER_SIZE)
dataloader = dataset.create_dataloader(batch_size=batch_size)
# Inference
y_prob = trainer.predict_step(dataloader)
y_pred = np.argmax(y_prob, axis=1)
label_encoder.decode(y_pred)
# Class distributions
prob_dist = get_probability_distribution(y_prob=y_prob[0], classes=label_encoder.classes)
print (json.dumps(prob_dist, indent=2))
import collections
import seaborn as sns
class InterpretableCNN(nn.Module):
def __init__(self, vocab_size, num_filters, filter_size,
hidden_dim, dropout_p, num_classes):
super(InterpretableCNN, self).__init__()
# Convolutional filters
self.filter_size = filter_size
self.conv = nn.Conv1d(
in_channels=vocab_size, out_channels=num_filters,
kernel_size=filter_size, stride=1, padding=0, padding_mode='zeros')
self.batch_norm = nn.BatchNorm1d(num_features=num_filters)
# FC layers
self.fc1 = nn.Linear(num_filters, hidden_dim)
self.dropout = nn.Dropout(dropout_p)
self.fc2 = nn.Linear(hidden_dim, num_classes)
def forward(self, inputs, channel_first=False, apply_softmax=False):
# Rearrange input so num_channels is in dim 1 (N, C, L)
x_in, = inputs
if not channel_first:
x_in = x_in.transpose(1, 2)
# Padding for `SAME` padding
max_seq_len = x_in.shape[2]
padding_left = int((self.conv.stride[0]*(max_seq_len-1) - max_seq_len + self.filter_size)/2)
padding_right = int(math.ceil((self.conv.stride[0]*(max_seq_len-1) - max_seq_len + self.filter_size)/2))
# Conv outputs
z = self.conv(F.pad(x_in, (padding_left, padding_right)))
return z
# Initialize
interpretable_model = InterpretableCNN(
vocab_size=len(tokenizer), num_filters=NUM_FILTERS, filter_size=FILTER_SIZE,
hidden_dim=HIDDEN_DIM, dropout_p=DROPOUT_P, num_classes=NUM_CLASSES)
# Load weights (same architecture)
interpretable_model.load_state_dict(torch.load(Path(dir, 'model.pt'), map_location=device))
interpretable_model.to(device)
# Initialize trainer
interpretable_trainer = Trainer(model=interpretable_model, device=device)
# Get conv outputs
conv_outputs = interpretable_trainer.predict_step(dataloader)
print (conv_outputs.shape) # (num_filters, max_seq_len)
# Visualize a bi-gram filter's outputs
tokens = tokenizer.sequences_to_texts(sequences)[0].split(' ')
sns.heatmap(conv_outputs, xticklabels=tokens)
| 0.847306 | 0.98987 |
# MAT 221 Calculus I
## April 9, 2020
Today's Agenda:
1. Continuous Function
2. Intermediate Value Theorem
3. Exercises
# Limits
A function limit, roughly speaking, describes the behavior of a function around a certain value. Limits play a role in the definition of the derivative and function continuity and are also used in the convergent sequences.
Before getting to the precise definition of a limit, we can investigate limit of a functon by plotting it and examining the area around the limit value.
For example, consider the limit (taken from James Stewart's *Calculus Early Transcendentals*, Exercise 11, page 107)
$$ \lim_{x\to 2} \frac{x^2 - x - 6}{x - 2} $$
We can plot this limit using the [`matplotlib.pylot`](https://matplotlib.org/api/pyplot_summary.html) module. The [numpy](http://www.numpy.org/) library is also imported for some convenience functions.
```
import numpy as np
import matplotlib.pyplot as plt
def f(x):
return (x ** 2 + x - 6) / (x - 2)
```
Get a set of values for $x$ using the [`linspace`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.linspace.html) function.
```
xvals = np.linspace(0, 2, 100, False)
```
Then plot the function using `matplotlib`.
```
plt.plot(xvals, f(xvals))
plt.show()
```
The plot shows that as $x$ gets closer to $2$, the function gets close to $5$. One way to verify this is to look closer at the values of the function around the limit.
```
xvals2 = np.linspace(1.90, 2, 100, False)
f(xvals2)
```
We can also use SymPy's [`limit`](http://docs.sympy.org/latest/tutorial/calculus.html#limits) function to calculate the limit.
```
from sympy import symbols, limit, sin, cos, init_printing
x = symbols('x')
init_printing()
limit((x ** 2 + x - 6) / (x - 2), x, 2)
```
Using this information, we can construct a more precise definition of a limit.
## Definition of a Limit
Limits are typically denoted as:
$$ \lim_{x\to a} \space f(x) = L $$
Or, alternatively:
$$ f(x) \rightarrow L, \qquad x \rightarrow a $$
In plain language, we can state the limit as, "the limit of the a function $f(x)$ as $x$ approaches $a$ is equal to $L$. For example, if we were considering the limit:
$$ \lim_{x \to 2} \space f(x) = 5 $$
We can state it as, "the limit of the function $f(x)$ as $x$ approaches $2$ is equal to $5$.
## One-Sided Limits
One-sided limits are used to express a limit as it approaches $a$ from a particular direction. The notation is similar to the limit seen above but with a slight change to indicate which direction $x$ is headed.
$$ \lim_{x \to a^+} \space f(x) = L, \qquad \lim_{x \to a^-} \space f(x) = L $$
The notation $x \to a^+$ states we are only interested in values of $x$ that are greater than $a$. Similarly, the notation $x \to a^-$ denotes our desire to our investigate values of $x$ less than $a$. These one-sided limits are also referred to as the "right-hand limit" and "left-hand limit", respectively.
For example, consider the function:
$$ \lim_{x \to -3^+} \space \frac{x + 2}{x + 3} $$
The $-3^+$ notation tells us we are only interested values greater than -3, thus the limit is a right-hand limit.
The function is not defined for $x = 3$, therefore we are dealing with an infinite limit. We can see the behavior of the function as $x$ approaches $-3$ by plotting.
```
def f2(x):
return (x + 2) / (x + 3)
xvals2 = np.linspace(-2, -3, 100, False)
xvals3 = np.linspace(-2.5, -3, 3000, False)
plt.figure(1, figsize=(14,3))
plt.subplot(121)
plt.plot(xvals2, f2(xvals2))
plt.subplot(122)
plt.plot(xvals3, f2(xvals3))
plt.xlim((-3.005, -2.99))
plt.show()
```
Both graphs approach $x = -3$ from the right and we can see the function quickly drop off as it gets closer to its limit. The graph on the right is a zoomed in representation of the graph on the left to better illustrate the infinite limit. Therefore, the limit of the function is $-\infty$, which we can confirm with SymPy.
```
limit((x + 2) / (x + 3), x, -3)
```
## References
Stewart, J. (2007). Essential calculus: Early transcendentals. Belmont, CA: Thomson Higher Education.
Strang, G. (2010). Calculus. Wellesley, MA: Wellesley-Cambridge.
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
def f(x):
return (x ** 2 + x - 6) / (x - 2)
xvals = np.linspace(0, 2, 100, False)
plt.plot(xvals, f(xvals))
plt.show()
xvals2 = np.linspace(1.90, 2, 100, False)
f(xvals2)
from sympy import symbols, limit, sin, cos, init_printing
x = symbols('x')
init_printing()
limit((x ** 2 + x - 6) / (x - 2), x, 2)
def f2(x):
return (x + 2) / (x + 3)
xvals2 = np.linspace(-2, -3, 100, False)
xvals3 = np.linspace(-2.5, -3, 3000, False)
plt.figure(1, figsize=(14,3))
plt.subplot(121)
plt.plot(xvals2, f2(xvals2))
plt.subplot(122)
plt.plot(xvals3, f2(xvals3))
plt.xlim((-3.005, -2.99))
plt.show()
limit((x + 2) / (x + 3), x, -3)
| 0.467818 | 0.99432 |
```
# !wget https://huggingface.co/datasets/huseinzol05/Malay-TTS-Yasmin/resolve/main/tts-malay-yasmin.tar.gz
# !wget https://huggingface.co/datasets/huseinzol05/Malay-TTS-Yasmin/resolve/main/normalized-texts.json
# !tar -xf tts-malay-yasmin.tar.gz
# !rm tts-malay-yasmin.tar.gz
# !wget https://huggingface.co/datasets/huseinzol05/Malay-TTS-Yasmin/resolve/main/tts-malay-yasmin-parliament.tar.gz
# !wget https://huggingface.co/datasets/huseinzol05/Malay-TTS-Yasmin/resolve/main/normalized-parliaments.json
# !tar -xf tts-malay-yasmin-parliament.tar.gz
# !rm tts-malay-yasmin-parliament.tar.gz
import os
import malaya_speech
from malaya_speech import Pipeline
from tqdm import tqdm
import numpy as np
import soundfile as sf
from glob import glob
config = {'sampling_rate': 22050,
'fft_size': 1024,
'hop_size': 256,
'win_length': None,
'window': 'hann',
'num_mels': 80,
'fmin': 0,
'fmax': None,
'global_gain_scale': 1.0,
'trim_silence': True}
directory = 'output-yasmin'
os.system(f'mkdir {directory}')
directories = ['audios']
for d in directories:
os.system(f'mkdir {directory}/{d}')
import json
with open('normalized-texts.json') as fopen:
texts = json.load(fopen)
with open('normalized-parliaments.json') as fopen:
parliament = json.load(fopen)
def process(txts,
start_silent_trail = int(0.05 * config['sampling_rate']),
middle_silent_trail = int(0.12 * config['sampling_rate']),
end_silent_trail = int(0.1 * config['sampling_rate']),
process_middle_silent = True):
txts = txts[0]
vad = malaya_speech.vad.webrtc()
for f in txts:
directory = f[2]
index = f[1]
f = f[0]
audio, _ = malaya_speech.load(f, sr = config['sampling_rate'])
audio = audio[start_silent_trail:]
if config['trim_silence']:
y_= malaya_speech.resample(audio, config['sampling_rate'], 16000)
y_ = malaya_speech.astype.float_to_int(y_)
frames = list(malaya_speech.generator.frames(audio, 30, config['sampling_rate']))
frames_ = list(malaya_speech.generator.frames(y_, 30, 16000, append_ending_trail = False))
frames_webrtc = [(frames[no], vad(frame)) for no, frame in enumerate(frames_)]
grouped_deep = malaya_speech.group.group_frames(frames_webrtc)
grouped_deep = malaya_speech.group.group_frames_threshold(grouped_deep, 0.15)
r = []
for no, g in enumerate(grouped_deep):
if g[1]:
g = g[0].array
else:
if no == 0:
g = g[0].array[-start_silent_trail:]
elif no == (len(grouped_deep) - 1):
g = g[0].array[:end_silent_trail]
else:
if process_middle_silent:
g = np.concatenate([g[0].array[:middle_silent_trail], g[0].array[-middle_silent_trail:]])
else:
g = g[0].array
r.append(g)
audio = np.concatenate(r)
sf.write(f'{directory}/audios/{index}.wav', audio, config['sampling_rate'])
txts = [(f"female/{t['index']}.wav", t['index'], directory) for t in texts]
i = 1508
process((txts[i: i + 10], 0))
from glob import glob
wavs = glob(f'{directory}/audios/*.wav')
wavs[:10]
import IPython.display as ipd
ipd.Audio(wavs[0])
import mp
for i in tqdm(range(0, len(txts), 1000)):
index = min(i + 1000, len(txts))
b = txts[i: index]
mp.multiprocessing(b, process, cores = 15, returned = False)
len(glob(f'{directory}/audios/*.wav'))
directory = 'output-yasmin-parliament'
os.system(f'mkdir {directory}')
directories = ['audios']
for d in directories:
os.system(f'mkdir {directory}/{d}')
txts = [(f"female-parliament/{t['index']}.wav", t['index'], directory) for t in parliament]
import mp
for i in tqdm(range(0, len(txts), 1000)):
index = min(i + 1000, len(txts))
b = txts[i: index]
mp.multiprocessing(b, process, cores = 15, returned = False)
directory = 'output-yasmin'
wavs = glob(f'/home/husein/speech-bahasa/{directory}/audios/*.wav')
yasmin = []
for f in wavs:
left = f
index = int(os.path.split(f)[1].replace('.wav', ''))
right = texts[index]['normalized']
yasmin.append((left, right))
yasmin[:2]
parliament_dict = {i['index']: i for i in parliament}
directory = 'output-yasmin-parliament'
wavs = glob(f'/home/husein/speech-bahasa/{directory}/audios/*.wav')
for f in wavs:
left = f
try:
index = int(os.path.split(f)[1].replace('.wav', ''))
right = parliament_dict[index]['normalized']
yasmin.append((left, right))
except Exception as e:
print(e)
len(yasmin)
from sklearn.model_selection import train_test_split
yasmin_train, yasmin_test = train_test_split(yasmin, test_size = 2000)
with open('yasmin-vits-test-set.txt', 'w') as fopen:
json.dump(yasmin_test, fopen)
with open('yasmin-vits-train-set.txt', 'w') as fopen:
json.dump(yasmin_train, fopen)
```
|
github_jupyter
|
# !wget https://huggingface.co/datasets/huseinzol05/Malay-TTS-Yasmin/resolve/main/tts-malay-yasmin.tar.gz
# !wget https://huggingface.co/datasets/huseinzol05/Malay-TTS-Yasmin/resolve/main/normalized-texts.json
# !tar -xf tts-malay-yasmin.tar.gz
# !rm tts-malay-yasmin.tar.gz
# !wget https://huggingface.co/datasets/huseinzol05/Malay-TTS-Yasmin/resolve/main/tts-malay-yasmin-parliament.tar.gz
# !wget https://huggingface.co/datasets/huseinzol05/Malay-TTS-Yasmin/resolve/main/normalized-parliaments.json
# !tar -xf tts-malay-yasmin-parliament.tar.gz
# !rm tts-malay-yasmin-parliament.tar.gz
import os
import malaya_speech
from malaya_speech import Pipeline
from tqdm import tqdm
import numpy as np
import soundfile as sf
from glob import glob
config = {'sampling_rate': 22050,
'fft_size': 1024,
'hop_size': 256,
'win_length': None,
'window': 'hann',
'num_mels': 80,
'fmin': 0,
'fmax': None,
'global_gain_scale': 1.0,
'trim_silence': True}
directory = 'output-yasmin'
os.system(f'mkdir {directory}')
directories = ['audios']
for d in directories:
os.system(f'mkdir {directory}/{d}')
import json
with open('normalized-texts.json') as fopen:
texts = json.load(fopen)
with open('normalized-parliaments.json') as fopen:
parliament = json.load(fopen)
def process(txts,
start_silent_trail = int(0.05 * config['sampling_rate']),
middle_silent_trail = int(0.12 * config['sampling_rate']),
end_silent_trail = int(0.1 * config['sampling_rate']),
process_middle_silent = True):
txts = txts[0]
vad = malaya_speech.vad.webrtc()
for f in txts:
directory = f[2]
index = f[1]
f = f[0]
audio, _ = malaya_speech.load(f, sr = config['sampling_rate'])
audio = audio[start_silent_trail:]
if config['trim_silence']:
y_= malaya_speech.resample(audio, config['sampling_rate'], 16000)
y_ = malaya_speech.astype.float_to_int(y_)
frames = list(malaya_speech.generator.frames(audio, 30, config['sampling_rate']))
frames_ = list(malaya_speech.generator.frames(y_, 30, 16000, append_ending_trail = False))
frames_webrtc = [(frames[no], vad(frame)) for no, frame in enumerate(frames_)]
grouped_deep = malaya_speech.group.group_frames(frames_webrtc)
grouped_deep = malaya_speech.group.group_frames_threshold(grouped_deep, 0.15)
r = []
for no, g in enumerate(grouped_deep):
if g[1]:
g = g[0].array
else:
if no == 0:
g = g[0].array[-start_silent_trail:]
elif no == (len(grouped_deep) - 1):
g = g[0].array[:end_silent_trail]
else:
if process_middle_silent:
g = np.concatenate([g[0].array[:middle_silent_trail], g[0].array[-middle_silent_trail:]])
else:
g = g[0].array
r.append(g)
audio = np.concatenate(r)
sf.write(f'{directory}/audios/{index}.wav', audio, config['sampling_rate'])
txts = [(f"female/{t['index']}.wav", t['index'], directory) for t in texts]
i = 1508
process((txts[i: i + 10], 0))
from glob import glob
wavs = glob(f'{directory}/audios/*.wav')
wavs[:10]
import IPython.display as ipd
ipd.Audio(wavs[0])
import mp
for i in tqdm(range(0, len(txts), 1000)):
index = min(i + 1000, len(txts))
b = txts[i: index]
mp.multiprocessing(b, process, cores = 15, returned = False)
len(glob(f'{directory}/audios/*.wav'))
directory = 'output-yasmin-parliament'
os.system(f'mkdir {directory}')
directories = ['audios']
for d in directories:
os.system(f'mkdir {directory}/{d}')
txts = [(f"female-parliament/{t['index']}.wav", t['index'], directory) for t in parliament]
import mp
for i in tqdm(range(0, len(txts), 1000)):
index = min(i + 1000, len(txts))
b = txts[i: index]
mp.multiprocessing(b, process, cores = 15, returned = False)
directory = 'output-yasmin'
wavs = glob(f'/home/husein/speech-bahasa/{directory}/audios/*.wav')
yasmin = []
for f in wavs:
left = f
index = int(os.path.split(f)[1].replace('.wav', ''))
right = texts[index]['normalized']
yasmin.append((left, right))
yasmin[:2]
parliament_dict = {i['index']: i for i in parliament}
directory = 'output-yasmin-parliament'
wavs = glob(f'/home/husein/speech-bahasa/{directory}/audios/*.wav')
for f in wavs:
left = f
try:
index = int(os.path.split(f)[1].replace('.wav', ''))
right = parliament_dict[index]['normalized']
yasmin.append((left, right))
except Exception as e:
print(e)
len(yasmin)
from sklearn.model_selection import train_test_split
yasmin_train, yasmin_test = train_test_split(yasmin, test_size = 2000)
with open('yasmin-vits-test-set.txt', 'w') as fopen:
json.dump(yasmin_test, fopen)
with open('yasmin-vits-train-set.txt', 'w') as fopen:
json.dump(yasmin_train, fopen)
| 0.305801 | 0.134634 |
```
import pandas as pd
import numpy as np
import os
from collections import defaultdict
from datetime import *
import sys
from pathlib import Path
sys.path.append(str(Path(os.getcwd()).parent.parent.absolute()))
try:
from lib_o_.o_fn import *
from lib_o_.o_rpadef import *
from lib_o_.o_rpafn import *
import lib_o_.o_time as oT
except:
from o_fn import *
from o_rpadef import *
from o_rpafn import *
import o_time as oT
def o_print(my_dict):
for key in my_dict.items():
x = my_dict.get(key)
def getvalue(my_dict, ky):
if ky is not None:
for key, value in my_dict.items():
if key in str (ky):
return value
else:
return 0
def codecorr(code,akey):
cd = code
if 'UNKNOW' in code:
for i in range(len(lss)):
vl = akey.find(lss[i])
if vl > 0 and vl is not None:
cd = akey[vl:vl+7]
break
else:
return cd
else:
return cd
def msgprep_head_znwise(hd = "Periodic Notification"):
nw = datetime.now()
dt = nw.strftime("%d-%m-%Y")
tm = nw.strftime("%H:%M")
a1 = hd + " at " + tm + " on " + dt
return a1
def catmap(df,omdb):
try:
dfdb = omdb
except:
dfdb = pd.read_csv(os.getcwd() + "\\csv_o_\\OMDB.csv")
df0 = df.rename (columns=str.upper)
ls = ['RESOURCE','CUSTOMATTR15','SUMMARY','ALERTKEY','LASTOCCURRENCE','CLEARTIMESTAMP']
df1 = df0[ls]
df1 = df1.assign(CAT = df1.apply (lambda x: TS (x.SUMMARY), axis=1))
df1 = df1.assign(CODE = df1.apply (lambda x: codecorr(x.CUSTOMATTR15, x.ALERTKEY), axis=1))
df2 = df1.assign(sCode = df1.apply (lambda x: x.CODE[0:5] if (x.CODE is not None) else "XXXXXXXX", axis=1))
df3 = df2.merge (dfdb, on='sCode')
df3['CODECAT'] = df3['CUSTOMATTR15'].str.cat(df3['CAT'])
df3['ZNCAT'] = df3['sZone'].str.cat(df3['CAT'])
df3 = df3.assign(CDLO = df3.apply (lambda x: x['CUSTOMATTR15'] + ": " + x['LASTOCCURRENCE'], axis=1))
try:
dfp1p2 = pd.read_csv(os.getcwd() + "\\csv_o_\\OMDB.csv")
df4 = df3.merge (dfp1p2, on='CUSTOMATTR15')
return df4
except:
return df3
def inner_list_to_dic(dic):
for key, value in dic.items():
dic[key] = set(value)
return dic
def joinls(l1,l2):
if len(l1)!=0 and len(l2)!=0:
lss = []
for i in l1:
for j in l2:
lss.append(str(i) + '$' + str(j))
return lss
elif len(l1)!=0 and len(l2)==0:
return l1
elif len(l1)==0 and len(l2)!=0:
return l2
else:
return l1
def colchk(df):
mcols = ['EQUIPMENTKEY','SITECODE','SUMMARY','ALERTKEY','LASTOCCURRENCE','CLEARTIMESTAMP']
ocols = ['RESOURCE','CUSTOMATTR15','SUMMARY','ALERTKEY','LASTOCCURRENCE','CLEARTIMESTAMP']
df = df.rename (columns=str.upper)
cols = df.columns.to_list()
if cols.count('SITECODE') != 0:
df = df.rename(columns={'SITECODE':'CUSTOMATTR15'})
if cols.count('EQUIPMENTKEY') != 0:
df = df.rename(columns={'EQUIPMENTKEY':'RESOURCE'})
for i in ocols:
if ocols.count(i) == 0:
print('must have column needs in table: but missing !',chr(10),mcols,chr(10),'exiting .....')
exit(0)
else:
sx = chrstream()
omnm(sx)
print(chr(10))
return df
def nested_dic_add(dc, k, v):
#map based on child dictionary and add new key value/append by key matching on clild
if len(dc)>0:
ln = len(list(dc))
for i in dc:
if k in list(dc[i]):
dc[i][k] = v if not list(dc[i]) else dc[i].get(k, []) + [v]
return dc
else:
if isinstance(v, list):
dc[ln+1] = {k:v}
else:
v1 = [v]
dc[ln+1] = {k:v1}
return dc
else:
if isinstance(v, list):
dc[1] = {k:v}
else:
v1 = [v]
dc[1] = {k:v1}
return dc
def mprep(df, dic, oncol):
heap = ""
gdc = defaultdict(dict)
for k, v in dic.items():
dff = df[df[k].isin(v)]
dff = dff.reset_index()
for i in range(len(v)):
hp = str(k) + "|" + str(dff.shape[0]) + chr(10) + dff[oncol].str.contains(v[i]).cat(sep=chr(10))
if heap == "":
heap = hp
else:
heap = heap + chr(10) + chr(10) + hp
else:
print(heap)
class o_rpa:
def __init__(self, mdf):
self.odb = omdb()
self.df0 = colchk(mdf)
self.df1 = mapdata(self.df0, self.odb)
self.df = self.df1
self.pickcols = "CDLO"
self.msgthread = defaultdict(dict)
self.mth = []
self.cond = defaultdict(dict)
self.lsky = []
self.lsvl = []
self.lsx = []
print(self.df.columns)
def regionwise(self, Name=[]):
self.msgthread = list(zn_dic())
def techwise(self):
self.msgthread = {'CAT': {'2G':"","3G":"","4G":""}}
def msgcol(self, lscols):
fault = 0
col = self.df.columns.to_list()
for i in lscols:
if col.count(i)==0:
fault = 1
else:
if fault == 0:
self.df = self.df.assign(pk = self.df[lscols].apply(lambda x: '- '.join(x.values.astype(str)), axis=1))
self.pickcols = "pk"
else:
print("!!!!!!!!!!!!!!!!! column name not found",chr(10),self.df.columns)
def csvmap(self, csvpath, match_col_name, column_to_pick = []):
try:
try:
dff = pd.read_csv(csvpath)
except:
dff = csvpath
if column_to_pick is not None:
dff = dff[column_to_pick]
ndf = self.df
self.df = ndf.merge (dff, on=match_col_name)
except:
print('csvpath not found')
def pnt(self):
print(self.df)
def getdf(self, current=True):
if current==False:
return self.df0
else:
return self.df
def sample(self):
print(self.df.head(5))
def summary(self):
print("COLUMN:", self.df.columns, chr(10))
print("Current Row: ", self.df.shape[0],'--',"Row Main:", self.df1.shape[0], chr(10))
print("filtering conditions : ", dict((k, v) for k, v in self.cond.items()))
print("msgformat by pickcols: ", 'CUSTOMATTR15','LASTOCCURRENCE')
print("msgthread: ", self.msgthread)
def condition(self,colname,colval):
ndc = nested_dic_add(self.cond,colname,colval)
self.cond = ndc.copy()
def rwise(self, whichzn=False):
if whichzn == False:
whichzn = "ALL"
if len(self.cond) == 1:
colsMain = list(self.cond[1])
rval = parsing(self.df, whichzn,[],colsMain[0], self.cond[1].get(colsMain[0]), False, False)
elif len(self.cond) == 2:
colsMain = list(self.cond[1])
cl = list(self.cond[2])
rval = parsing(self.df, whichzn,self.pickcols,colsMain[0], self.cond[1].get(colsMain[0]), cl[0],
self.cond[2].get(cl[0]))
else:
print('under development')
def regionwise_count(self, list_cat):
rv = zonewise_count(self.df1, list_cat)
print('----------------------------------------')
return rv
def update_cond_1(self, ky, vl):
self.cond.setdefault(ky, []).append(vl)
def update_cond_2(self, ky, vl):
if isinstance(vl,list):
self.cond[ky] = vl if not list(self.cond) else self.cond.get(ky, []) + vl
else:
vlx = [vl]
self.cond[ky] = vlx if not list(self.cond) else self.cond.get(ky, []) + vlx
def timecal(self, start_time_colname, end_time_colname=False):
self.df = oT.dfdiff(self.df, start_time_colname, end_time_colname)
print("new column name is 'DUR'")
def timefmt(self, colname, fmt="%Y/%m/%d %H:%M:%S"):
if len(fmt) <= 9:
self.df[colname] = self.df.apply(lambda x: oT.sec_to_dur(x[colname]), axis = 1)
print(self.df[colname])
else:
self.df[colname] = oT.datetime_convert_format(self.df, colname, fmt)
print(self.df[colname])
def add2(self, k, v):
self.df = self.df[self.df[k].isin(v)]
self.lsky.append(k)
self.lsvl.append(v)
xk = joinls(self.lsx,v)
self.lsx = xk
print(self.lsx)
try:
self.df = self.df[self.df[k].isin(v)]
except:
print('except trigger')
def gen3(self):
hpf = chr(10)
ndf = self.df
print(ndf.columns)
for i in range(len(self.lsx)):
spl = self.lsx[i].split('$')
if len(self.lsky) == len(spl):
xdf = self.df
for n in range(len(spl)):
k = self.lsky
v = spl[n]
xdf = xdf[xdf[k[n]].isin([v])]
else:
x11 = " | ".join(spl) + ": " + str(xdf.shape[0])
if xdf.shape[0] != 0:
x11 = x11 + chr(10) + xdf[self.pickcols].str.cat(sep=chr(10))
print(x11, chr(10))
def pntcond(self):
L = list(self.cond.values())
Lx = []
if (len(L[0]))<=1:
L1 = [item for sublist in L[0] for item in sublist]
Lx.append(L1)
else:
Lx.append(L1)
print(os.getcwd())
df = pd.read_csv(os.getcwd() + "\\sclick.csv") # data source,
xx = o_rpa(df)
xx.timecal('LASTOCCURRENCE')
xx.timefmt('DUR','%H:%M:%S')
xx.update_cond_1('sZone',['COM','NOA'])
xx.update_cond_1('CAT',['2G','3G','4G'])
xx.update_cond_1('P1P2',['P1','P2'])
xx.sample()
xx.msgcol(['CUSTOMATTR15','LASTOCCURRENCE','P1P2'])
xx.pntcond()
#xx.gen3()
#rval = xx.rwise()
#ST = xx.regionwise_count(['2G','3G','4G','MF','DL'])
#print(ST)
#dc = {'sZone':{0:{'COM':{'CAT':{0:'2G'}}}}}
#print(dc)
#slicedf(dx,dc)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import os
from collections import defaultdict
from datetime import *
import sys
from pathlib import Path
sys.path.append(str(Path(os.getcwd()).parent.parent.absolute()))
try:
from lib_o_.o_fn import *
from lib_o_.o_rpadef import *
from lib_o_.o_rpafn import *
import lib_o_.o_time as oT
except:
from o_fn import *
from o_rpadef import *
from o_rpafn import *
import o_time as oT
def o_print(my_dict):
for key in my_dict.items():
x = my_dict.get(key)
def getvalue(my_dict, ky):
if ky is not None:
for key, value in my_dict.items():
if key in str (ky):
return value
else:
return 0
def codecorr(code,akey):
cd = code
if 'UNKNOW' in code:
for i in range(len(lss)):
vl = akey.find(lss[i])
if vl > 0 and vl is not None:
cd = akey[vl:vl+7]
break
else:
return cd
else:
return cd
def msgprep_head_znwise(hd = "Periodic Notification"):
nw = datetime.now()
dt = nw.strftime("%d-%m-%Y")
tm = nw.strftime("%H:%M")
a1 = hd + " at " + tm + " on " + dt
return a1
def catmap(df,omdb):
try:
dfdb = omdb
except:
dfdb = pd.read_csv(os.getcwd() + "\\csv_o_\\OMDB.csv")
df0 = df.rename (columns=str.upper)
ls = ['RESOURCE','CUSTOMATTR15','SUMMARY','ALERTKEY','LASTOCCURRENCE','CLEARTIMESTAMP']
df1 = df0[ls]
df1 = df1.assign(CAT = df1.apply (lambda x: TS (x.SUMMARY), axis=1))
df1 = df1.assign(CODE = df1.apply (lambda x: codecorr(x.CUSTOMATTR15, x.ALERTKEY), axis=1))
df2 = df1.assign(sCode = df1.apply (lambda x: x.CODE[0:5] if (x.CODE is not None) else "XXXXXXXX", axis=1))
df3 = df2.merge (dfdb, on='sCode')
df3['CODECAT'] = df3['CUSTOMATTR15'].str.cat(df3['CAT'])
df3['ZNCAT'] = df3['sZone'].str.cat(df3['CAT'])
df3 = df3.assign(CDLO = df3.apply (lambda x: x['CUSTOMATTR15'] + ": " + x['LASTOCCURRENCE'], axis=1))
try:
dfp1p2 = pd.read_csv(os.getcwd() + "\\csv_o_\\OMDB.csv")
df4 = df3.merge (dfp1p2, on='CUSTOMATTR15')
return df4
except:
return df3
def inner_list_to_dic(dic):
for key, value in dic.items():
dic[key] = set(value)
return dic
def joinls(l1,l2):
if len(l1)!=0 and len(l2)!=0:
lss = []
for i in l1:
for j in l2:
lss.append(str(i) + '$' + str(j))
return lss
elif len(l1)!=0 and len(l2)==0:
return l1
elif len(l1)==0 and len(l2)!=0:
return l2
else:
return l1
def colchk(df):
mcols = ['EQUIPMENTKEY','SITECODE','SUMMARY','ALERTKEY','LASTOCCURRENCE','CLEARTIMESTAMP']
ocols = ['RESOURCE','CUSTOMATTR15','SUMMARY','ALERTKEY','LASTOCCURRENCE','CLEARTIMESTAMP']
df = df.rename (columns=str.upper)
cols = df.columns.to_list()
if cols.count('SITECODE') != 0:
df = df.rename(columns={'SITECODE':'CUSTOMATTR15'})
if cols.count('EQUIPMENTKEY') != 0:
df = df.rename(columns={'EQUIPMENTKEY':'RESOURCE'})
for i in ocols:
if ocols.count(i) == 0:
print('must have column needs in table: but missing !',chr(10),mcols,chr(10),'exiting .....')
exit(0)
else:
sx = chrstream()
omnm(sx)
print(chr(10))
return df
def nested_dic_add(dc, k, v):
#map based on child dictionary and add new key value/append by key matching on clild
if len(dc)>0:
ln = len(list(dc))
for i in dc:
if k in list(dc[i]):
dc[i][k] = v if not list(dc[i]) else dc[i].get(k, []) + [v]
return dc
else:
if isinstance(v, list):
dc[ln+1] = {k:v}
else:
v1 = [v]
dc[ln+1] = {k:v1}
return dc
else:
if isinstance(v, list):
dc[1] = {k:v}
else:
v1 = [v]
dc[1] = {k:v1}
return dc
def mprep(df, dic, oncol):
heap = ""
gdc = defaultdict(dict)
for k, v in dic.items():
dff = df[df[k].isin(v)]
dff = dff.reset_index()
for i in range(len(v)):
hp = str(k) + "|" + str(dff.shape[0]) + chr(10) + dff[oncol].str.contains(v[i]).cat(sep=chr(10))
if heap == "":
heap = hp
else:
heap = heap + chr(10) + chr(10) + hp
else:
print(heap)
class o_rpa:
def __init__(self, mdf):
self.odb = omdb()
self.df0 = colchk(mdf)
self.df1 = mapdata(self.df0, self.odb)
self.df = self.df1
self.pickcols = "CDLO"
self.msgthread = defaultdict(dict)
self.mth = []
self.cond = defaultdict(dict)
self.lsky = []
self.lsvl = []
self.lsx = []
print(self.df.columns)
def regionwise(self, Name=[]):
self.msgthread = list(zn_dic())
def techwise(self):
self.msgthread = {'CAT': {'2G':"","3G":"","4G":""}}
def msgcol(self, lscols):
fault = 0
col = self.df.columns.to_list()
for i in lscols:
if col.count(i)==0:
fault = 1
else:
if fault == 0:
self.df = self.df.assign(pk = self.df[lscols].apply(lambda x: '- '.join(x.values.astype(str)), axis=1))
self.pickcols = "pk"
else:
print("!!!!!!!!!!!!!!!!! column name not found",chr(10),self.df.columns)
def csvmap(self, csvpath, match_col_name, column_to_pick = []):
try:
try:
dff = pd.read_csv(csvpath)
except:
dff = csvpath
if column_to_pick is not None:
dff = dff[column_to_pick]
ndf = self.df
self.df = ndf.merge (dff, on=match_col_name)
except:
print('csvpath not found')
def pnt(self):
print(self.df)
def getdf(self, current=True):
if current==False:
return self.df0
else:
return self.df
def sample(self):
print(self.df.head(5))
def summary(self):
print("COLUMN:", self.df.columns, chr(10))
print("Current Row: ", self.df.shape[0],'--',"Row Main:", self.df1.shape[0], chr(10))
print("filtering conditions : ", dict((k, v) for k, v in self.cond.items()))
print("msgformat by pickcols: ", 'CUSTOMATTR15','LASTOCCURRENCE')
print("msgthread: ", self.msgthread)
def condition(self,colname,colval):
ndc = nested_dic_add(self.cond,colname,colval)
self.cond = ndc.copy()
def rwise(self, whichzn=False):
if whichzn == False:
whichzn = "ALL"
if len(self.cond) == 1:
colsMain = list(self.cond[1])
rval = parsing(self.df, whichzn,[],colsMain[0], self.cond[1].get(colsMain[0]), False, False)
elif len(self.cond) == 2:
colsMain = list(self.cond[1])
cl = list(self.cond[2])
rval = parsing(self.df, whichzn,self.pickcols,colsMain[0], self.cond[1].get(colsMain[0]), cl[0],
self.cond[2].get(cl[0]))
else:
print('under development')
def regionwise_count(self, list_cat):
rv = zonewise_count(self.df1, list_cat)
print('----------------------------------------')
return rv
def update_cond_1(self, ky, vl):
self.cond.setdefault(ky, []).append(vl)
def update_cond_2(self, ky, vl):
if isinstance(vl,list):
self.cond[ky] = vl if not list(self.cond) else self.cond.get(ky, []) + vl
else:
vlx = [vl]
self.cond[ky] = vlx if not list(self.cond) else self.cond.get(ky, []) + vlx
def timecal(self, start_time_colname, end_time_colname=False):
self.df = oT.dfdiff(self.df, start_time_colname, end_time_colname)
print("new column name is 'DUR'")
def timefmt(self, colname, fmt="%Y/%m/%d %H:%M:%S"):
if len(fmt) <= 9:
self.df[colname] = self.df.apply(lambda x: oT.sec_to_dur(x[colname]), axis = 1)
print(self.df[colname])
else:
self.df[colname] = oT.datetime_convert_format(self.df, colname, fmt)
print(self.df[colname])
def add2(self, k, v):
self.df = self.df[self.df[k].isin(v)]
self.lsky.append(k)
self.lsvl.append(v)
xk = joinls(self.lsx,v)
self.lsx = xk
print(self.lsx)
try:
self.df = self.df[self.df[k].isin(v)]
except:
print('except trigger')
def gen3(self):
hpf = chr(10)
ndf = self.df
print(ndf.columns)
for i in range(len(self.lsx)):
spl = self.lsx[i].split('$')
if len(self.lsky) == len(spl):
xdf = self.df
for n in range(len(spl)):
k = self.lsky
v = spl[n]
xdf = xdf[xdf[k[n]].isin([v])]
else:
x11 = " | ".join(spl) + ": " + str(xdf.shape[0])
if xdf.shape[0] != 0:
x11 = x11 + chr(10) + xdf[self.pickcols].str.cat(sep=chr(10))
print(x11, chr(10))
def pntcond(self):
L = list(self.cond.values())
Lx = []
if (len(L[0]))<=1:
L1 = [item for sublist in L[0] for item in sublist]
Lx.append(L1)
else:
Lx.append(L1)
print(os.getcwd())
df = pd.read_csv(os.getcwd() + "\\sclick.csv") # data source,
xx = o_rpa(df)
xx.timecal('LASTOCCURRENCE')
xx.timefmt('DUR','%H:%M:%S')
xx.update_cond_1('sZone',['COM','NOA'])
xx.update_cond_1('CAT',['2G','3G','4G'])
xx.update_cond_1('P1P2',['P1','P2'])
xx.sample()
xx.msgcol(['CUSTOMATTR15','LASTOCCURRENCE','P1P2'])
xx.pntcond()
#xx.gen3()
#rval = xx.rwise()
#ST = xx.regionwise_count(['2G','3G','4G','MF','DL'])
#print(ST)
#dc = {'sZone':{0:{'COM':{'CAT':{0:'2G'}}}}}
#print(dc)
#slicedf(dx,dc)
| 0.095856 | 0.162646 |
```
import os
import numpy as np
import pandas as pd
import seaborn as sns
sns.set(rc={'figure.figsize':(10, 10)})
def load_metrics(input_dir, method="MoFA"):
dirname = os.path.join(input_dir, method)
if not os.path.exists(dirname):
return None
metrics_file = os.path.join(dirname, "metrics.npy")
if not os.path.exists(metrics_file):
print("Metrics file does not exist for method {}".format(method))
return None
else:
metrics = np.load(metrics_file, allow_pickle=True)
metrics = metrics.item()
return metrics
def get_dataframe_to_plot(base_dir, mining_dir):
data = dict()
for subtype in [2, 3, 4, 5, 6]:
methods_dir = os.path.join(base_dir, "num_subtypes_{}".format(subtype))
for method in os.listdir(methods_dir):
metrics = load_metrics(input_dir=methods_dir, method=method)
if metrics is not None:
for metric, value in metrics.items():
if metric == "mean-runtime":
continue
if metric not in data:
num_vals = len(value)
data[metric] = value
else:
num_vals = len(value)
data[metric] += value
if "method" not in data:
data["method"] = [method] * num_vals
else:
data["method"] += [method] * num_vals
metrics = load_metrics(input_dir=mining_dir, method="Mining")
if metrics is not None:
for metric, value in metrics.items():
if metric == "mean-runtime":
continue
if metric not in data:
num_vals = len(value)
data[metric] = value
else:
num_vals = len(value)
data[metric] += value
if "method" not in data:
data["method"] = ["Mining"] * num_vals
else:
data["method"] += ["Mining"] * num_vals
return pd.DataFrame(data)
OUT_DIR = "../outputs/"
GNW_OUTPUTS_DIR = "../outputs/gnw/"
INTERSIM_SAMPLES = 100
GNW_SAMPLES = 25
GNW_ENTITIES = 500
NUM_SUBTYPES = 3
#mode = "intersim"
mode = "gnw"
GNW_EXAMPLE_DIR = os.path.join(GNW_OUTPUTS_DIR, "{}_samples_{}_genes".format(GNW_SAMPLES, GNW_ENTITIES))
INTERSIM_DIR = os.path.join(OUT_DIR, "intersim_" + str(INTERSIM_SAMPLES) + "_samples")
if mode == "intersim":
BASE_DIR = INTERSIM_DIR
else:
BASE_DIR = GNW_EXAMPLE_DIR
METHODS_DIR = os.path.join(BASE_DIR, "num_subtypes_" + str(NUM_SUBTYPES))
MINING_DIR = BASE_DIR
df = get_dataframe_to_plot(BASE_DIR, MINING_DIR)
print(df.columns)
methods = df.method.unique()
metric = "avg_sil_width"
if metric == "avg_sil_width":
y_label = "Silhouette_score"
elif metric == "corrected_rand":
y_label = "Adjusted Rand Index"
ax = sns.boxplot(x="method", y=metric, data=df, order=methods)
if mode == "gnw":
title = "GeneNetWeaver: SAMPLES: {}, GENES: {}".format(GNW_SAMPLES, GNW_ENTITIES, NUM_SUBTYPES)
else:
title = "InterSIM: SAMPLES {}".format(INTERSIM_SAMPLES, NUM_SUBTYPES)
ax.set_title(title)
ax.set_ylabel(y_label)
ax.set_xlabel("Method used")
#ax.set_ylim(0,)
ax1 = sns.lineplot(x="method", y=metric, data=df)
ax1.set_ylabel("Silhouette Score")
if mode == "gnw":
title = "GeneNetWeaver: SAMPLES: {}, GENES: {} SUBTYPES: {}".format(GNW_SAMPLES, GNW_ENTITIES, NUM_SUBTYPES)
else:
title = "InterSIM: SAMPLES {}, SUBTYPES: {}".format(INTERSIM_SAMPLES, NUM_SUBTYPES)
ax1.set_title(title)
ax.set_ylim(0, 0.5)
mri = sns.load_dataset("fmri")
mri.head()
```
|
github_jupyter
|
import os
import numpy as np
import pandas as pd
import seaborn as sns
sns.set(rc={'figure.figsize':(10, 10)})
def load_metrics(input_dir, method="MoFA"):
dirname = os.path.join(input_dir, method)
if not os.path.exists(dirname):
return None
metrics_file = os.path.join(dirname, "metrics.npy")
if not os.path.exists(metrics_file):
print("Metrics file does not exist for method {}".format(method))
return None
else:
metrics = np.load(metrics_file, allow_pickle=True)
metrics = metrics.item()
return metrics
def get_dataframe_to_plot(base_dir, mining_dir):
data = dict()
for subtype in [2, 3, 4, 5, 6]:
methods_dir = os.path.join(base_dir, "num_subtypes_{}".format(subtype))
for method in os.listdir(methods_dir):
metrics = load_metrics(input_dir=methods_dir, method=method)
if metrics is not None:
for metric, value in metrics.items():
if metric == "mean-runtime":
continue
if metric not in data:
num_vals = len(value)
data[metric] = value
else:
num_vals = len(value)
data[metric] += value
if "method" not in data:
data["method"] = [method] * num_vals
else:
data["method"] += [method] * num_vals
metrics = load_metrics(input_dir=mining_dir, method="Mining")
if metrics is not None:
for metric, value in metrics.items():
if metric == "mean-runtime":
continue
if metric not in data:
num_vals = len(value)
data[metric] = value
else:
num_vals = len(value)
data[metric] += value
if "method" not in data:
data["method"] = ["Mining"] * num_vals
else:
data["method"] += ["Mining"] * num_vals
return pd.DataFrame(data)
OUT_DIR = "../outputs/"
GNW_OUTPUTS_DIR = "../outputs/gnw/"
INTERSIM_SAMPLES = 100
GNW_SAMPLES = 25
GNW_ENTITIES = 500
NUM_SUBTYPES = 3
#mode = "intersim"
mode = "gnw"
GNW_EXAMPLE_DIR = os.path.join(GNW_OUTPUTS_DIR, "{}_samples_{}_genes".format(GNW_SAMPLES, GNW_ENTITIES))
INTERSIM_DIR = os.path.join(OUT_DIR, "intersim_" + str(INTERSIM_SAMPLES) + "_samples")
if mode == "intersim":
BASE_DIR = INTERSIM_DIR
else:
BASE_DIR = GNW_EXAMPLE_DIR
METHODS_DIR = os.path.join(BASE_DIR, "num_subtypes_" + str(NUM_SUBTYPES))
MINING_DIR = BASE_DIR
df = get_dataframe_to_plot(BASE_DIR, MINING_DIR)
print(df.columns)
methods = df.method.unique()
metric = "avg_sil_width"
if metric == "avg_sil_width":
y_label = "Silhouette_score"
elif metric == "corrected_rand":
y_label = "Adjusted Rand Index"
ax = sns.boxplot(x="method", y=metric, data=df, order=methods)
if mode == "gnw":
title = "GeneNetWeaver: SAMPLES: {}, GENES: {}".format(GNW_SAMPLES, GNW_ENTITIES, NUM_SUBTYPES)
else:
title = "InterSIM: SAMPLES {}".format(INTERSIM_SAMPLES, NUM_SUBTYPES)
ax.set_title(title)
ax.set_ylabel(y_label)
ax.set_xlabel("Method used")
#ax.set_ylim(0,)
ax1 = sns.lineplot(x="method", y=metric, data=df)
ax1.set_ylabel("Silhouette Score")
if mode == "gnw":
title = "GeneNetWeaver: SAMPLES: {}, GENES: {} SUBTYPES: {}".format(GNW_SAMPLES, GNW_ENTITIES, NUM_SUBTYPES)
else:
title = "InterSIM: SAMPLES {}, SUBTYPES: {}".format(INTERSIM_SAMPLES, NUM_SUBTYPES)
ax1.set_title(title)
ax.set_ylim(0, 0.5)
mri = sns.load_dataset("fmri")
mri.head()
| 0.315525 | 0.162181 |
# PageRank
In this notebook, we will use both NetworkX and cuGraph to compute the PageRank of each vertex in our test dataset. The NetworkX and cuGraph processes will be interleaved so that each step can be compared.
Notebook Credits
* Original Authors: Bradley Rees and James Wyles
* Created: 08/13/2019
* Updated: 04/06/2022
RAPIDS Versions: 22.04
Test Hardware
* GV100 32G, CUDA 11.5
## Introduction
Pagerank is measure of the relative importance, also called centrality, of a vertex based on the relative importance of it's neighbors. PageRank was developed by Google and is (was) used to rank it's search results. PageRank uses the connectivity information of a graph to rank the importance of each vertex.
See [Wikipedia](https://en.wikipedia.org/wiki/PageRank) for more details on the algorithm.
To compute the Pagerank scores for a graph in cuGraph we use:<br>
**cugraph.pagerank(G,alpha=0.85, max_iter=100, tol=1.0e-5)**
* __G__: cugraph.Graph object
* __alpha__: float, The damping factor represents the probability to follow an outgoing edge. default is 0.85
* __max_iter__: int, The maximum number of iterations before an answer is returned. This can be used to limit the execution time and do an early exit before the solver reaches the convergence tolerance. If this value is lower or equal to 0 cuGraph will use the default value, which is 100
* __tol__: float, Set the tolerance the approximation, this parameter should be a small magnitude value. The lower the tolerance the better the approximation. If this value is 0.0f, cuGraph will use the default value which is 0.00001. Setting too small a tolerance can lead to non-convergence due to numerical roundoff. Usually values between 0.01 and 0.00001 are acceptable.
Returns:
* __df__: a cudf.DataFrame object with two columns:
* df['vertex']: The vertex identifier for the vertex
* df['pagerank']: The pagerank score for the vertex
### Some notes about vertex IDs...
* The current version of cuGraph requires that vertex IDs be representable as 32-bit integers, meaning graphs currently can contain at most 2^32 unique vertex IDs. However, this limitation is being actively addressed and a version of cuGraph that accommodates more than 2^32 vertices will be available in the near future.
* cuGraph will automatically renumber graphs to an internal format consisting of a contiguous series of integers starting from 0, and convert back to the original IDs when returning data to the caller. If the vertex IDs of the data are already a contiguous series of integers starting from 0, the auto-renumbering step can be skipped for faster graph creation times.
* To skip auto-renumbering, set the `renumber` boolean arg to `False` when calling the appropriate graph creation API (eg. `G.from_cudf_edgelist(gdf_r, source='src', destination='dst', renumber=False)`).
* For more advanced renumbering support, see the examples in `structure/renumber.ipynb` and `structure/renumber-2.ipynb`
### Test Data
We will be using the Zachary Karate club dataset
*W. W. Zachary, An information flow model for conflict and fission in small groups, Journal of
Anthropological Research 33, 452-473 (1977).*

### Prep
```
# The notebook compares cuGraph to NetworkX,
# therefore there some additional non-RAPIDS python libraries need to be installed.
# Please run this cell if you need the additional libraries
!pip install networkx
!pip install scipy
# Import needed libraries
import cugraph
import cudf
from collections import OrderedDict
# NetworkX libraries
import networkx as nx
from scipy.io import mmread
```
### Some Prep
```
# define the parameters
max_iter = 100 # The maximum number of iterations
tol = 0.00001 # tolerance
alpha = 0.85 # alpha
# Define the path to the test data
datafile='../data/karate-data.csv'
```
---
# NetworkX
```
# Read the data, this also created a NetworkX Graph
file = open(datafile, 'rb')
Gnx = nx.read_edgelist(file)
pr_nx = nx.pagerank(Gnx, alpha=alpha, max_iter=max_iter, tol=tol)
pr_nx
```
Running NetworkX is that easy.
Let's seet how that compares to cuGraph
----
# cuGraph
### Read in the data - GPU
cuGraph graphs can be created from cuDF, dask_cuDF and Pandas dataframes
The data file contains an edge list, which represents the connection of a vertex to another. The `source` to `destination` pairs is in what is known as Coordinate Format (COO). In this test case, the data is just two columns. However a third, `weight`, column is also possible
```
# Read the data
gdf = cudf.read_csv(datafile, names=["src", "dst"], delimiter='\t', dtype=["int32", "int32"] )
```
### Create a Graph
```
# create a Graph using the source (src) and destination (dst) vertex pairs from the Dataframe
G = cugraph.from_edgelist(gdf, source='src', destination='dst')
```
### Call the PageRank algorithm
```
# Call cugraph.pagerank to get the pagerank scores
gdf_page = cugraph.pagerank(G)
```
_It was that easy!_
Compared to NetworkX, the cuGraph data loading might have been more steps, but using cuDF allows for a wider range of data to be loaded.
----
Let's now look at the results
```
# Find the most important vertex using the scores
# This methods should only be used for small graph
bestScore = gdf_page['pagerank'][0]
bestVert = gdf_page['vertex'][0]
for i in range(len(gdf_page)):
if gdf_page['pagerank'].iloc[i] > bestScore:
bestScore = gdf_page['pagerank'].iloc[i]
bestVert = gdf_page['vertex'].iloc[i]
print("Best vertex is " + str(bestVert) + " with score of " + str(bestScore))
```
The top PageRank vertex and socre match what was found by NetworkX
```
# A better way to do that would be to find the max and then use that values in a query
pr_max = gdf_page['pagerank'].max()
def print_pagerank_threshold(_df, t=0) :
filtered = _df.query('pagerank >= @t')
for i in range(len(filtered)):
print("Best vertex is " + str(filtered['vertex'].iloc[i]) +
" with score of " + str(filtered['pagerank'].iloc[i]))
print_pagerank_threshold(gdf_page, pr_max)
```
----
a PageRank score of _0.10047_ is very low, which can be an indication that there is no more central vertex than any other. Rather than just looking at the top score, let's look at the top three vertices and see if there are any insights that can be inferred.
Since this is a very small graph, let's just sort and get the first three records
```
sort_pr = gdf_page.sort_values('pagerank', ascending=False)
sort_pr.head(3)
```
Going back and looking at the graph with the top three vertices highlighted (illustration below) it is easy to see that the top scoring vertices also appear to be the vertices with the most connections.
Let's look at sorted list of degrees (since the graph is undirected and symmetrized, the out degree is the same as the in degree)
```
d = G.degrees()
d.sort_values('out_degree', ascending=False).head(3)
```
<img src="../img/zachary_graph_pagerank.png" width="600">
----
# Personalized PageRank
The issue with PageRank is that it sets the initial weights of all the nodes the same. In other words, it assumes a uniform starting probability for every node. What if we have a priori information about the nodes? We can use personalized PageRank (PPR) to assist.
```
# Let's bump up some weights and see how that changes the results
personalization_vec = cudf.DataFrame()
personalization_vec['vertex'] = [17, 26]
personalization_vec['values'] = [0.5, 0.75]
ppr = cugraph.pagerank(G, alpha=0.85, personalization=personalization_vec, max_iter=100, tol=1.0e-5, nstart=None)
ppr.sort_values('pagerank', ascending=False).head(3)
# looking at the initial PageRank values
gdf_page[gdf_page['vertex'].isin([26,17,1])]
```
___
Copyright (c) 2019-2020, NVIDIA CORPORATION.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
___
|
github_jupyter
|
# The notebook compares cuGraph to NetworkX,
# therefore there some additional non-RAPIDS python libraries need to be installed.
# Please run this cell if you need the additional libraries
!pip install networkx
!pip install scipy
# Import needed libraries
import cugraph
import cudf
from collections import OrderedDict
# NetworkX libraries
import networkx as nx
from scipy.io import mmread
# define the parameters
max_iter = 100 # The maximum number of iterations
tol = 0.00001 # tolerance
alpha = 0.85 # alpha
# Define the path to the test data
datafile='../data/karate-data.csv'
# Read the data, this also created a NetworkX Graph
file = open(datafile, 'rb')
Gnx = nx.read_edgelist(file)
pr_nx = nx.pagerank(Gnx, alpha=alpha, max_iter=max_iter, tol=tol)
pr_nx
# Read the data
gdf = cudf.read_csv(datafile, names=["src", "dst"], delimiter='\t', dtype=["int32", "int32"] )
# create a Graph using the source (src) and destination (dst) vertex pairs from the Dataframe
G = cugraph.from_edgelist(gdf, source='src', destination='dst')
# Call cugraph.pagerank to get the pagerank scores
gdf_page = cugraph.pagerank(G)
# Find the most important vertex using the scores
# This methods should only be used for small graph
bestScore = gdf_page['pagerank'][0]
bestVert = gdf_page['vertex'][0]
for i in range(len(gdf_page)):
if gdf_page['pagerank'].iloc[i] > bestScore:
bestScore = gdf_page['pagerank'].iloc[i]
bestVert = gdf_page['vertex'].iloc[i]
print("Best vertex is " + str(bestVert) + " with score of " + str(bestScore))
# A better way to do that would be to find the max and then use that values in a query
pr_max = gdf_page['pagerank'].max()
def print_pagerank_threshold(_df, t=0) :
filtered = _df.query('pagerank >= @t')
for i in range(len(filtered)):
print("Best vertex is " + str(filtered['vertex'].iloc[i]) +
" with score of " + str(filtered['pagerank'].iloc[i]))
print_pagerank_threshold(gdf_page, pr_max)
sort_pr = gdf_page.sort_values('pagerank', ascending=False)
sort_pr.head(3)
d = G.degrees()
d.sort_values('out_degree', ascending=False).head(3)
# Let's bump up some weights and see how that changes the results
personalization_vec = cudf.DataFrame()
personalization_vec['vertex'] = [17, 26]
personalization_vec['values'] = [0.5, 0.75]
ppr = cugraph.pagerank(G, alpha=0.85, personalization=personalization_vec, max_iter=100, tol=1.0e-5, nstart=None)
ppr.sort_values('pagerank', ascending=False).head(3)
# looking at the initial PageRank values
gdf_page[gdf_page['vertex'].isin([26,17,1])]
| 0.611034 | 0.987277 |
# Amazon SageMaker PyTorch コンテナを用いた MNIST の学習と推論
## 目次
1. [背景](#1.背景)
1. [セットアップ](#2.セットアップ)
1. [データ](#3.データ)
1. [学習](#4.学習)
1. [ハイパーパラメータ調整を用いた学習](#5.ハイパーパラメータ調整を用いた学習)
1. [ホスティング](#6.ホスティング)
1. [リソースの削除](#7.リソースの削除)
---
## 1.背景
MNISTは、手書き文字の分類に広く使用されているデータセットです。 70,000個のラベル付きの28x28ピクセルの手書き数字のグレースケール画像で構成されています。 データセットは、60,000個のトレーニング画像と10,000個のテスト画像に分割されます。 手書きの数字 0から9の合計10のクラスがあります。 このチュートリアルでは、PyTorch を使用して SageMaker で MNIST モデルをトレーニングおよびテストする方法を示します。 また、SageMaker の自動モデルチューニングを使用して適切なハイパーパラメーターを選択し、最適なモデルを取得する方法をご説明します。
SageMaker の PyTorch の詳細については、[sagemaker-pytorch-containers](https://github.com/aws/sagemaker-pytorch-containers) と [sagemaker-python-sdk](https://github.com/aws/sagemaker-python-sdk) のレポジトリをご参照ください。
---
## 2.セットアップ
SageMaker セッションを作成し、設定を開始します。
- 学習およびモデルデータに使用する S3 バケットとプレフィックスは、ノートブックインスタンス、トレーニング、およびホスティングと同じリージョン内にある必要があります。
- データへの学習およびホスティングアクセスを提供するために使用される IAM ロール arn を用います。 ノートブックインスタンス、学習インスタンス、および/またはホスティングインスタンスに複数のロールが必要な場合は、 `sagemaker.get_execution_role()` を、適切な IAM ロール arn 文字列に置き換えてください。
```
import sagemaker
from sagemaker.tuner import IntegerParameter, CategoricalParameter, ContinuousParameter, HyperparameterTuner
sagemaker_session = sagemaker.Session()
bucket = sagemaker_session.default_bucket()
prefix = 'sagemaker/DEMO-pytorch-mnist'
role = sagemaker.get_execution_role()
```
このノートブックのコードは、以前からのノートブックインスタンスで実行する場合と、SageMaker Studio のノートブックで実行する場合とで挙動が異なります。以下のセルを実行することで、いまの実行環境が以前からのノートブックインスタンスなのか、SageMaker Studio のノートブックなのかを判定して、`on_studio`に記録します。この結果に基づいて、以降のノートブックの実行を次のように変更します。
- データセットの展開先を変更します。SageMaker Studio を利用する場合、home のディレクトリは EFS をマウントして実現されており、データセットを展開する際にやや時間を要します。そこで home 以外のところへ展開するようにします。
- SageMaker Studio では学習・推論の local モードがサポートされていません。従って、Studio では local モードを行わないようにします。
```
import os, json
NOTEBOOK_METADATA_FILE = "/opt/ml/metadata/resource-metadata.json"
if os.path.exists(NOTEBOOK_METADATA_FILE):
with open(NOTEBOOK_METADATA_FILE, "rb") as f:
metadata = json.loads(f.read())
domain_id = metadata.get("DomainId")
on_studio = True if domain_id is not None else False
print("Is this notebook runnning on Studio?: {}".format(on_studio))
```
## 3.データ
### 3.1.データの取得
```
!aws s3 cp s3://fast-ai-imageclas/mnist_png.tgz . --no-sign-request
if on_studio:
!tar -xzf mnist_png.tgz -C /opt/ml --no-same-owner
else:
!tar -xvzf mnist_png.tgz
from torchvision import datasets, transforms
from torch.utils.data import DataLoader
import torch
import os
root_dir_studio = '/opt/ml'
data_dir = os.path.join(root_dir_studio,'data') if on_studio else 'data'
training_dir = os.path.join(root_dir_studio,'mnist_png/training') if on_studio else 'mnist_png/training'
test_dir = os.path.join(root_dir_studio,'mnist_png/testing') if on_studio else 'mnist_png/testing'
os.makedirs(data_dir, exist_ok=True)
training_data = datasets.ImageFolder(root=training_dir,
transform=transforms.Compose([
transforms.Grayscale(),
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))]))
test_data = datasets.ImageFolder(root=test_dir,
transform=transforms.Compose([
transforms.Grayscale(),
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))]))
training_data_loader = DataLoader(training_data, batch_size=len(training_data))
training_data_loaded = next(iter(training_data_loader))
torch.save(training_data_loaded, os.path.join(data_dir, 'training.pt'))
test_data_loader = DataLoader(test_data, batch_size=len(test_data))
test_data_loaded = next(iter(test_data_loader))
torch.save(test_data_loaded, os.path.join(data_dir, 'test.pt'))
```
### 3.2.データをS3にアップロードする
データセットを S3 にアップロードするには、 `sagemaker.Session.upload_data` 関数を使用します。 戻り値として入力した S3 のロケーションは、後で学習ジョブを実行するときに使用します。
```
inputs = sagemaker_session.upload_data(path=data_dir, bucket=bucket, key_prefix=prefix)
print('input spec (in this case, just an S3 path): {}'.format(inputs))
```
## 4.学習
### 4.1.学習スクリプト
`mnist.py` スクリプトは、SageMaker モデルの学習とホスティングに必要なすべてのコードを提供します(`model_fn` はモデルをロードするための関数です)。
学習スクリプトは、SageMaker の外部で実行するトレーニングスクリプトに非常に似ていますが、次のようなさまざまな環境変数を通じてトレーニング環境に関する有用なプロパティにアクセスできます。
* `SM_MODEL_DIR`: モデルアーティファクトを書き込むディレクトリへのパスを表す文字列。モデルは、推論用ホスティングのためにS3にアップロードされます。
* `SM_NUM_GPUS`: 現在のコンテナで利用可能な GPU の数。
* `SM_CURRENT_HOST`: コンテナネットワーク上の現在のコンテナの名前。
* `SM_HOSTS`: すべてのホストを含む JSON エンコードリスト。
`fit()`メソッドの呼び出しで1つの入力チャンネル `training` が使用されたとすると、 `SM_CHANNEL_ [channel_name]`の形式に従って以下が設定されます:
* `SM_CHANNEL_TRAINING`:`training` チャンネルのデータを含むディレクトリへのパスを表す文字列。
学習に関する環境変数の詳細については、[SageMaker Containers](https://github.com/aws/sagemaker-containers) をご覧ください。
典型的なトレーニングスクリプトの書き方は下記の通りです。入力チャンネルからデータをロードし、ハイパーパラメーターで学習の設定、モデルを学習、モデルを `model_dir` に保存し、そこから学習済みモデルのホスティングを行います。 ハイパーパラメーターは引数としてスクリプトに渡され、 `argparse.ArgumentParser` インスタンスとして取得できます。
下記は、このノートブックで使われるスクリプト ` mnist.py` です。SageMaker PyTorch Container で定義されるデフォルトの実装方法に従い [sagemaker-pytorch-containers](https://github.com/aws/sagemaker-pytorch-containers) 、 `input_fn` 、` predict_fn` 、 `output_fn` および ` transform_fn` の実装を使用します。メインガード (``if __name__=='__main__':``) の中のスクリプトは、学習時にのみ実行され、モデルのホスティング時には実行されません。上記のように、必要な `mnist.py` スクリプトに ` model_fn` を実装しています。
```
!pygmentize mnist.py
```
### 4.2.Estimatorの定義
学習の条件を設定するため、Estimator クラスの子クラスの PyTorch オブジェクトを作成します。 ここでは、PyTorchスクリプト、IAMロール、および(ジョブごとの)ハードウェア構成を渡す PyTorch Estimator を定義しています。また合わせてローカルの `source_dir` を指定することで、依存するスクリプト群をコンテナにコピーして、学習時に使用することが可能です。
`mnist.py` で定義されているハイパーパラメータをレンジの形で指定してハイパーパラメータ探索を行う場合は、[ハイパーパラメーター調整ジョブをセットアップする](### ハイパーパラメーター調整ジョブをセットアップする) で紹介しています。
```
from sagemaker.pytorch import PyTorch
if on_studio:
instance_type = 'ml.m4.xlarge'
else:
instance_type = 'local'
estimator = PyTorch(entry_point="mnist.py",
role=role,
framework_version='1.8.0',
py_version='py3',
instance_count=1,
instance_type=instance_type,
hyperparameters={
'batch-size':128,
'lr': 0.01,
'epochs': 1,
'backend': 'gloo'
})
```
### 4.3.ローカルモードでの学習の実行
***以前のノートブックインスタンスのみローカルモードを利用できます。Studio の場合は ml.m4.xlarge を利用します。***
`fit()` メソッドで学習ジョブを実行します。`entry_point` で指定したローカルのスクリプトが、学習用のコンテナ内で実行されます。
ノートブックインスタンスのCPUで学習する場合はinstance_type = 'local'、GPUで学習する場合はlocal_gpuを指定します。インスタンス数は、ノートブックインスタンスの数、すなわち1になるため、 train_instance_countに指定された値が1より大きい場合も1として扱われますが、traini_instance_count > 1 の分散学習はローカル環境でも擬似的に検証できます。
```
estimator.fit({'training': inputs})
```
### 4.4.ローカルモードでモデルの検証
***以前のノートブックインスタンスのみローカルモードを利用できます。Studio の場合は ml.m4.xlarge を利用します。***
```
predictor = estimator.deploy(initial_instance_count=1, instance_type=instance_type)
```
### 4.5.画像データに対する予測の実行
正規化していない MNIST データを正解データとして確認するために読み込んでおきます。テストデータから5枚をランダムにサンプリングして予測してみます。予測のために`predictor`に画像を入力します。
```
%matplotlib inline
import random
import numpy as np
import matplotlib.pyplot as plt
raw_test_data = datasets.ImageFolder(root=test_dir,
transform=transforms.Compose([
transforms.Grayscale(),
transforms.ToTensor()]))
num_samples = 5
indices = random.sample(range(len(raw_test_data) - 1), num_samples)
raw_images = np.array([raw_test_data[i][0].numpy() for i in indices])
raw_labels = np.array([raw_test_data[i][1] for i in indices])
images = np.array([test_data[i][0].numpy() for i in indices])
for i in range(num_samples):
plt.subplot(1,num_samples,i+1)
plt.imshow(raw_images[i].reshape(28, 28), cmap='gray')
plt.title(raw_labels[i])
plt.axis('off')
prediction = predictor.predict(images)
predicted_label = prediction.argmax(axis=1)
print('The predicted labels are: {}'.format(predicted_label))
```
## 5 ハイパーパラメータ調整を用いた学習
### 5.1.ハイパーパラメーター調整ジョブの Estimator を設定
*以下のデフォルト設定では、ハイパーパラメーター調整ジョブが完了するまでに10分程度かかります。*
この例では、 SageMaker Python SDK を使用して、ハイパーパラメーターの最適化を取り入れた学習を行います。先ほどはノートブックインスタンス上で学習を行うのローカルモードを扱いましたが、ここでは、`ml.m4.xlarge` を学習インスタンスとして個別に起動し、その上にPyTorchコンテナを起動し、学習する方法を説明します。
ハイパーパラメータ調整とは、ディープラーニングでよく用いられるパラメータの最適化の手法です。SageMaker は、学習率、バッチサイズ、エポック数などのパラメータの最適値を探索するためのインターフェースを持っています。探索したいハイパーパラメーターごとに、連続値として探索する場合はその範囲、またはリストの中から探索したい場合は探索可能な値のリストを指定します。ハイパーパラメーター調整ジョブは、異なるハイパーパラメーター設定で複数のトレーニングジョブを並列に起動し、定義済みの `objective metric` (目的関数) に基づいてそれらのトレーニングジョブの結果を評価します。以前の結果に基づいて次のハイパーパラメーター探索の設定を選択します。ハイパーパラメーター調整ジョブごとに、一度に並列で実行する学習インスタンス数、最大の学習ジョブ数、をそれぞれ割り当て、最大の学習ジョブ数が実行されると探索を終え、`objective metric` に対して最適なパフォーマンスを達成したハイパーパラメータの組み合わせを返します。
次に、以下の手順に従って、SageMaker Python SDK を使用してハイパーパラメーター調整ジョブをセットアップします。
* `estimator` を作成して PyTorch 学習ジョブをセットアップします。
* 調整するハイパーパラメーターの範囲を定義します。この例では、学習率とバッチサイズのハイパーパラメータ探索範囲を設定します。
* 最適化するチューニングジョブの `objective metric` を定義します。
* 上記の設定で `HyperparameterTuner` オブジェクトを設定します。
Spot Instanceを用いて実行する場合は、下記のコードを Estimator の train_instance_type の次の行に追加しましょう。
```python
max_run = 5000,
use_spot_instances = 'True',
max_wait = 10000,
```
```
from sagemaker.pytorch import PyTorch
hpo_estimator = PyTorch(entry_point="mnist.py",
role=role,
framework_version='1.8.0',
py_version='py3',
instance_count=1,
instance_type='ml.m4.xlarge',
hyperparameters={
'epochs': 4,
'backend': 'gloo'
})
```
`Estimator` を定義したら、調整するハイパーパラメーターとその探索範囲を指定します。ハイパーパラメーターの探索範囲の指定は3種類の方法があります。
- カテゴリーパラメーターは、探索したい値のリストを `CategoricalParameter(list)` で定義します。このリストの中から最適な値を探索します。
- 連続パラメーターは、`ContinuousParameter(min、max)` で定義される最小値と最大値の間の連続空間で、任意の実数値から探索を行います。
- 整数パラメーターは、`IntegerParameter(min、max)` で定義された最小値と最大値の間の任意の整数値で、探索を行います。
* 可能であれば、値を最も制限の少ないタイプとして指定することをお勧めします。 たとえば、0.01〜0.2の連続値として学習率を調整すると、0.01、0.1、0.15、または0.2の値を持つカテゴリパラメーターとして調整するよりも良い結果が得られる可能性があります。 バッチサイズは一般的に2のべき乗であることが推奨されているため、ここではカテゴリパラメータとしてバッチサイズを指定しています。
```
hyperparameter_ranges = {'lr': ContinuousParameter(0.001, 0.1),'batch-size': CategoricalParameter([32,64,128,256])}
```
次に objective metric とその定義を指定します。これには、トレーニングジョブの CloudWatch ログからそのメトリックを抽出するために必要な正規表現(Regex)が含まれます。 この特定のケースでは、スクリプトは平均損失値を出力し、それを objective metric として使用します。また、`objective_type` を `minimize` に設定します。これにより、ハイパーパラメーターチューニングは、最適なハイパーパラメーター設定を検索するときに客観的なメトリックを最小化しようとします。 デフォルトでは、 `objective_type` は `maximize` に設定されています。
```
objective_metric_name = 'average test loss'
objective_type = 'Minimize'
metric_definitions = [{'Name': 'average test loss',
'Regex': 'Test set: Average loss: ([0-9\\.]+)'}]
```
### 5.2.ハイパーパラメーター調整ジョブの tuner 設定する
次に、 `HyperparameterTuner` オブジェクトを作成します。
- 上記で作成した PyTorch 推定器
- ハイパーパラメーターの範囲
- Objective metric 名と定義
- 合計で実行するトレーニングジョブの数や並行して実行できるトレーニングジョブの数などのリソース構成の調整。
```
tuner = HyperparameterTuner(hpo_estimator,
objective_metric_name,
hyperparameter_ranges,
metric_definitions,
max_jobs=4,
max_parallel_jobs=2,
objective_type=objective_type)
```
### 5.3.ハイパーパラメーター調整ジョブを起動する
`.fit()`を呼び出し、学習およびテストデータセットへの S3 パスを渡すことで、ハイパーパラメーターチューニングジョブを開始できます。
ハイパーパラメーターチューニングジョブが作成されたら、次のステップでチューニングジョブを記述してその進行状況を確認できます。SageMaker コンソールからジョブに移動して、ハイパーパラメーターチューニングジョブの進行状況を確認できます。
```
tuner.fit({'training': inputs})
tuner.wait()
```
## 6.ホスティング
### 6.1.エンドポイントを作成する
トレーニング完了後、Tuner オブジェクトを使用して、`PyTorchPredictor` をビルドおよびデプロイします。前の手順では、Tuner が複数のトレーニングジョブを起動し、最適な obejctive metric を持つ結果のモデルが最適なモデルとして定義ました。これにより、SageMaker エンドポイントが作成されます。これにより、チューナーが探索した最適なモデルに基づいて推論を実行するためのホスティングを行います。
deploy 関数の引数により、エンドポイントに使用されるインスタンスの数とタイプを設定できます。これらは、トレーニングジョブで使用した値と同じである必要はありません。ここでは、モデルを単一の ```ml.m4.xlarge``` インスタンスにデプロイします。
```
predictor_hpo = tuner.deploy(initial_instance_count=1, instance_type='ml.m4.xlarge')
```
### 6.2.評価
Estimator を使用して、手書きの数字を分類できるようになりました。Jupyer Notebook のみで利用可能です。 (Jupyter lab では利用できません)
下のセルを実行すると、空の画像ボックスが表示されます。 次に、その中に数字を描画すると、ピクセルデータがこのノートブックの `data` 変数にロードされ、`predictor` に渡すことができます。
```
from IPython.display import HTML
HTML(open("input.html").read())
```
### - HPOを用いた学習の結果
```
import numpy as np
image = np.array([data], dtype=np.float32)
response_hpo = predictor_hpo.predict(image)
prediction_hpo = response_hpo.argmax(axis=1)[0]
print(prediction_hpo)
```
### - ローカルモードの学習の結果
```
image = np.array([data], dtype=np.float32)
response = predictor.predict(image)
prediction = response.argmax(axis=1)[0]
print(prediction)
```
## 7.リソースの削除
上記で作成したホスティングエンドポイントは、明示的に削除しないと立ち上がったままになり、課金が継続されます。不要な課金を防ぐために、このノートブックの実行が終了したらエンドポイントを削除しましょう。立ち上がっている同一リージョン内のエンドポイントは、SageMaker マネージメントコンソールの「推論」->「エンドポイント」から一覧を確認できます。
```
# tuner で作ったエンドポイントの削除
predictor_hpo.delete_endpoint()
```
|
github_jupyter
|
import sagemaker
from sagemaker.tuner import IntegerParameter, CategoricalParameter, ContinuousParameter, HyperparameterTuner
sagemaker_session = sagemaker.Session()
bucket = sagemaker_session.default_bucket()
prefix = 'sagemaker/DEMO-pytorch-mnist'
role = sagemaker.get_execution_role()
import os, json
NOTEBOOK_METADATA_FILE = "/opt/ml/metadata/resource-metadata.json"
if os.path.exists(NOTEBOOK_METADATA_FILE):
with open(NOTEBOOK_METADATA_FILE, "rb") as f:
metadata = json.loads(f.read())
domain_id = metadata.get("DomainId")
on_studio = True if domain_id is not None else False
print("Is this notebook runnning on Studio?: {}".format(on_studio))
!aws s3 cp s3://fast-ai-imageclas/mnist_png.tgz . --no-sign-request
if on_studio:
!tar -xzf mnist_png.tgz -C /opt/ml --no-same-owner
else:
!tar -xvzf mnist_png.tgz
from torchvision import datasets, transforms
from torch.utils.data import DataLoader
import torch
import os
root_dir_studio = '/opt/ml'
data_dir = os.path.join(root_dir_studio,'data') if on_studio else 'data'
training_dir = os.path.join(root_dir_studio,'mnist_png/training') if on_studio else 'mnist_png/training'
test_dir = os.path.join(root_dir_studio,'mnist_png/testing') if on_studio else 'mnist_png/testing'
os.makedirs(data_dir, exist_ok=True)
training_data = datasets.ImageFolder(root=training_dir,
transform=transforms.Compose([
transforms.Grayscale(),
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))]))
test_data = datasets.ImageFolder(root=test_dir,
transform=transforms.Compose([
transforms.Grayscale(),
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))]))
training_data_loader = DataLoader(training_data, batch_size=len(training_data))
training_data_loaded = next(iter(training_data_loader))
torch.save(training_data_loaded, os.path.join(data_dir, 'training.pt'))
test_data_loader = DataLoader(test_data, batch_size=len(test_data))
test_data_loaded = next(iter(test_data_loader))
torch.save(test_data_loaded, os.path.join(data_dir, 'test.pt'))
inputs = sagemaker_session.upload_data(path=data_dir, bucket=bucket, key_prefix=prefix)
print('input spec (in this case, just an S3 path): {}'.format(inputs))
!pygmentize mnist.py
from sagemaker.pytorch import PyTorch
if on_studio:
instance_type = 'ml.m4.xlarge'
else:
instance_type = 'local'
estimator = PyTorch(entry_point="mnist.py",
role=role,
framework_version='1.8.0',
py_version='py3',
instance_count=1,
instance_type=instance_type,
hyperparameters={
'batch-size':128,
'lr': 0.01,
'epochs': 1,
'backend': 'gloo'
})
estimator.fit({'training': inputs})
predictor = estimator.deploy(initial_instance_count=1, instance_type=instance_type)
%matplotlib inline
import random
import numpy as np
import matplotlib.pyplot as plt
raw_test_data = datasets.ImageFolder(root=test_dir,
transform=transforms.Compose([
transforms.Grayscale(),
transforms.ToTensor()]))
num_samples = 5
indices = random.sample(range(len(raw_test_data) - 1), num_samples)
raw_images = np.array([raw_test_data[i][0].numpy() for i in indices])
raw_labels = np.array([raw_test_data[i][1] for i in indices])
images = np.array([test_data[i][0].numpy() for i in indices])
for i in range(num_samples):
plt.subplot(1,num_samples,i+1)
plt.imshow(raw_images[i].reshape(28, 28), cmap='gray')
plt.title(raw_labels[i])
plt.axis('off')
prediction = predictor.predict(images)
predicted_label = prediction.argmax(axis=1)
print('The predicted labels are: {}'.format(predicted_label))
max_run = 5000,
use_spot_instances = 'True',
max_wait = 10000,
from sagemaker.pytorch import PyTorch
hpo_estimator = PyTorch(entry_point="mnist.py",
role=role,
framework_version='1.8.0',
py_version='py3',
instance_count=1,
instance_type='ml.m4.xlarge',
hyperparameters={
'epochs': 4,
'backend': 'gloo'
})
hyperparameter_ranges = {'lr': ContinuousParameter(0.001, 0.1),'batch-size': CategoricalParameter([32,64,128,256])}
objective_metric_name = 'average test loss'
objective_type = 'Minimize'
metric_definitions = [{'Name': 'average test loss',
'Regex': 'Test set: Average loss: ([0-9\\.]+)'}]
tuner = HyperparameterTuner(hpo_estimator,
objective_metric_name,
hyperparameter_ranges,
metric_definitions,
max_jobs=4,
max_parallel_jobs=2,
objective_type=objective_type)
tuner.fit({'training': inputs})
tuner.wait()
predictor_hpo = tuner.deploy(initial_instance_count=1, instance_type='ml.m4.xlarge')
from IPython.display import HTML
HTML(open("input.html").read())
import numpy as np
image = np.array([data], dtype=np.float32)
response_hpo = predictor_hpo.predict(image)
prediction_hpo = response_hpo.argmax(axis=1)[0]
print(prediction_hpo)
image = np.array([data], dtype=np.float32)
response = predictor.predict(image)
prediction = response.argmax(axis=1)[0]
print(prediction)
# tuner で作ったエンドポイントの削除
predictor_hpo.delete_endpoint()
| 0.410402 | 0.953837 |
```
import math
import torch
import torch.nn as nn
import torch.nn.functional as F
class RNNModel(nn.Module):
"""Container module with an encoder, a recurrent module, and a decoder."""
def __init__(self, rnn_type, ntoken, ninp, nhid, nlayers, dropout=0.5, tie_weights=False):
super(RNNModel, self).__init__()
self.ntoken = ntoken
self.drop = nn.Dropout(dropout)
self.encoder = nn.Embedding(ntoken, ninp)
if rnn_type in ['LSTM', 'GRU']:
self.rnn = getattr(nn, rnn_type)(ninp, nhid, nlayers, dropout=dropout)
else:
try:
nonlinearity = {'RNN_TANH': 'tanh', 'RNN_RELU': 'relu'}[rnn_type]
except KeyError:
raise ValueError( """An invalid option for `--model` was supplied,
options are ['LSTM', 'GRU', 'RNN_TANH' or 'RNN_RELU']""")
self.rnn = nn.RNN(ninp, nhid, nlayers, nonlinearity=nonlinearity, dropout=dropout)
self.decoder = nn.Linear(nhid, ntoken)
# Optionally tie weights as in:
# "Using the Output Embedding to Improve Language Models" (Press & Wolf 2016)
# https://arxiv.org/abs/1608.05859
# and
# "Tying Word Vectors and Word Classifiers: A Loss Framework for Language Modeling" (Inan et al. 2016)
# https://arxiv.org/abs/1611.01462
if tie_weights:
if nhid != ninp:
raise ValueError('When using the tied flag, nhid must be equal to emsize')
self.decoder.weight = self.encoder.weight
self.init_weights()
self.rnn_type = rnn_type
self.nhid = nhid
self.nlayers = nlayers
def init_weights(self):
initrange = 0.1
nn.init.uniform_(self.encoder.weight, -initrange, initrange)
nn.init.zeros_(self.decoder.weight)
nn.init.uniform_(self.decoder.weight, -initrange, initrange)
def forward(self, input, hidden):
emb = self.drop(self.encoder(input))
output, hidden = self.rnn(emb, hidden)
output = self.drop(output)
decoded = self.decoder(output)
decoded = decoded.view(-1, self.ntoken)
return F.log_softmax(decoded, dim=1), hidden
def init_hidden(self, bsz):
weight = next(self.parameters())
if self.rnn_type == 'LSTM':
return (weight.new_zeros(self.nlayers, bsz, self.nhid),
weight.new_zeros(self.nlayers, bsz, self.nhid))
else:
return weight.new_zeros(self.nlayers, bsz, self.nhid)
# Temporarily leave PositionalEncoding module here. Will be moved somewhere else.
class PositionalEncoding(nn.Module):
r"""Inject some information about the relative or absolute position of the tokens
in the sequence. The positional encodings have the same dimension as
the embeddings, so that the two can be summed. Here, we use sine and cosine
functions of different frequencies.
.. math::
\text{PosEncoder}(pos, 2i) = sin(pos/10000^(2i/d_model))
\text{PosEncoder}(pos, 2i+1) = cos(pos/10000^(2i/d_model))
\text{where pos is the word position and i is the embed idx)
Args:
d_model: the embed dim (required).
dropout: the dropout value (default=0.1).
max_len: the max. length of the incoming sequence (default=5000).
Examples:
>>> pos_encoder = PositionalEncoding(d_model)
"""
def __init__(self, d_model, dropout=0.1, max_len=5000):
super(PositionalEncoding, self).__init__()
self.dropout = nn.Dropout(p=dropout)
pe = torch.zeros(max_len, d_model)
position = torch.arange(0, max_len, dtype=torch.float).unsqueeze(1)
div_term = torch.exp(torch.arange(0, d_model, 2).float() * (-math.log(10000.0) / d_model))
pe[:, 0::2] = torch.sin(position * div_term)
pe[:, 1::2] = torch.cos(position * div_term)
pe = pe.unsqueeze(0).transpose(0, 1)
self.register_buffer('pe', pe)
def forward(self, x):
r"""Inputs of forward function
Args:
x: the sequence fed to the positional encoder model (required).
Shape:
x: [sequence length, batch size, embed dim]
output: [sequence length, batch size, embed dim]
Examples:
>>> output = pos_encoder(x)
"""
x = x + self.pe[:x.size(0), :]
return self.dropout(x)
class TransformerModel(nn.Module):
"""Container module with an encoder, a recurrent or transformer module, and a decoder."""
def __init__(self, ntoken, ninp, nhead, nhid, nlayers, dropout=0.5):
super(TransformerModel, self).__init__()
try:
from torch.nn import TransformerEncoder, TransformerEncoderLayer
except:
raise ImportError('TransformerEncoder module does not exist in PyTorch 1.1 or lower.')
self.model_type = 'Transformer'
self.src_mask = None
self.pos_encoder = PositionalEncoding(ninp, dropout)
encoder_layers = TransformerEncoderLayer(ninp, nhead, nhid, dropout)
self.transformer_encoder = TransformerEncoder(encoder_layers, nlayers)
self.encoder = nn.Embedding(ntoken, ninp)
self.ninp = ninp
self.decoder = nn.Linear(ninp, ntoken)
self.init_weights()
def _generate_square_subsequent_mask(self, sz):
mask = (torch.triu(torch.ones(sz, sz)) == 1).transpose(0, 1)
mask = mask.float().masked_fill(mask == 0, float('-inf')).masked_fill(mask == 1, float(0.0))
return mask
def init_weights(self):
initrange = 0.1
nn.init.uniform_(self.encoder.weight, -initrange, initrange)
nn.init.zeros_(self.decoder.weight)
nn.init.uniform_(self.decoder.weight, -initrange, initrange)
def forward(self, src, has_mask=True):
if has_mask:
device = src.device
if self.src_mask is None or self.src_mask.size(0) != len(src):
mask = self._generate_square_subsequent_mask(len(src)).to(device)
self.src_mask = mask
else:
self.src_mask = None
src = self.encoder(src) * math.sqrt(self.ninp)
src = self.pos_encoder(src)
output = self.transformer_encoder(src, self.src_mask)
output = self.decoder(output)
return F.log_softmax(output, dim=-1)
```
|
github_jupyter
|
import math
import torch
import torch.nn as nn
import torch.nn.functional as F
class RNNModel(nn.Module):
"""Container module with an encoder, a recurrent module, and a decoder."""
def __init__(self, rnn_type, ntoken, ninp, nhid, nlayers, dropout=0.5, tie_weights=False):
super(RNNModel, self).__init__()
self.ntoken = ntoken
self.drop = nn.Dropout(dropout)
self.encoder = nn.Embedding(ntoken, ninp)
if rnn_type in ['LSTM', 'GRU']:
self.rnn = getattr(nn, rnn_type)(ninp, nhid, nlayers, dropout=dropout)
else:
try:
nonlinearity = {'RNN_TANH': 'tanh', 'RNN_RELU': 'relu'}[rnn_type]
except KeyError:
raise ValueError( """An invalid option for `--model` was supplied,
options are ['LSTM', 'GRU', 'RNN_TANH' or 'RNN_RELU']""")
self.rnn = nn.RNN(ninp, nhid, nlayers, nonlinearity=nonlinearity, dropout=dropout)
self.decoder = nn.Linear(nhid, ntoken)
# Optionally tie weights as in:
# "Using the Output Embedding to Improve Language Models" (Press & Wolf 2016)
# https://arxiv.org/abs/1608.05859
# and
# "Tying Word Vectors and Word Classifiers: A Loss Framework for Language Modeling" (Inan et al. 2016)
# https://arxiv.org/abs/1611.01462
if tie_weights:
if nhid != ninp:
raise ValueError('When using the tied flag, nhid must be equal to emsize')
self.decoder.weight = self.encoder.weight
self.init_weights()
self.rnn_type = rnn_type
self.nhid = nhid
self.nlayers = nlayers
def init_weights(self):
initrange = 0.1
nn.init.uniform_(self.encoder.weight, -initrange, initrange)
nn.init.zeros_(self.decoder.weight)
nn.init.uniform_(self.decoder.weight, -initrange, initrange)
def forward(self, input, hidden):
emb = self.drop(self.encoder(input))
output, hidden = self.rnn(emb, hidden)
output = self.drop(output)
decoded = self.decoder(output)
decoded = decoded.view(-1, self.ntoken)
return F.log_softmax(decoded, dim=1), hidden
def init_hidden(self, bsz):
weight = next(self.parameters())
if self.rnn_type == 'LSTM':
return (weight.new_zeros(self.nlayers, bsz, self.nhid),
weight.new_zeros(self.nlayers, bsz, self.nhid))
else:
return weight.new_zeros(self.nlayers, bsz, self.nhid)
# Temporarily leave PositionalEncoding module here. Will be moved somewhere else.
class PositionalEncoding(nn.Module):
r"""Inject some information about the relative or absolute position of the tokens
in the sequence. The positional encodings have the same dimension as
the embeddings, so that the two can be summed. Here, we use sine and cosine
functions of different frequencies.
.. math::
\text{PosEncoder}(pos, 2i) = sin(pos/10000^(2i/d_model))
\text{PosEncoder}(pos, 2i+1) = cos(pos/10000^(2i/d_model))
\text{where pos is the word position and i is the embed idx)
Args:
d_model: the embed dim (required).
dropout: the dropout value (default=0.1).
max_len: the max. length of the incoming sequence (default=5000).
Examples:
>>> pos_encoder = PositionalEncoding(d_model)
"""
def __init__(self, d_model, dropout=0.1, max_len=5000):
super(PositionalEncoding, self).__init__()
self.dropout = nn.Dropout(p=dropout)
pe = torch.zeros(max_len, d_model)
position = torch.arange(0, max_len, dtype=torch.float).unsqueeze(1)
div_term = torch.exp(torch.arange(0, d_model, 2).float() * (-math.log(10000.0) / d_model))
pe[:, 0::2] = torch.sin(position * div_term)
pe[:, 1::2] = torch.cos(position * div_term)
pe = pe.unsqueeze(0).transpose(0, 1)
self.register_buffer('pe', pe)
def forward(self, x):
r"""Inputs of forward function
Args:
x: the sequence fed to the positional encoder model (required).
Shape:
x: [sequence length, batch size, embed dim]
output: [sequence length, batch size, embed dim]
Examples:
>>> output = pos_encoder(x)
"""
x = x + self.pe[:x.size(0), :]
return self.dropout(x)
class TransformerModel(nn.Module):
"""Container module with an encoder, a recurrent or transformer module, and a decoder."""
def __init__(self, ntoken, ninp, nhead, nhid, nlayers, dropout=0.5):
super(TransformerModel, self).__init__()
try:
from torch.nn import TransformerEncoder, TransformerEncoderLayer
except:
raise ImportError('TransformerEncoder module does not exist in PyTorch 1.1 or lower.')
self.model_type = 'Transformer'
self.src_mask = None
self.pos_encoder = PositionalEncoding(ninp, dropout)
encoder_layers = TransformerEncoderLayer(ninp, nhead, nhid, dropout)
self.transformer_encoder = TransformerEncoder(encoder_layers, nlayers)
self.encoder = nn.Embedding(ntoken, ninp)
self.ninp = ninp
self.decoder = nn.Linear(ninp, ntoken)
self.init_weights()
def _generate_square_subsequent_mask(self, sz):
mask = (torch.triu(torch.ones(sz, sz)) == 1).transpose(0, 1)
mask = mask.float().masked_fill(mask == 0, float('-inf')).masked_fill(mask == 1, float(0.0))
return mask
def init_weights(self):
initrange = 0.1
nn.init.uniform_(self.encoder.weight, -initrange, initrange)
nn.init.zeros_(self.decoder.weight)
nn.init.uniform_(self.decoder.weight, -initrange, initrange)
def forward(self, src, has_mask=True):
if has_mask:
device = src.device
if self.src_mask is None or self.src_mask.size(0) != len(src):
mask = self._generate_square_subsequent_mask(len(src)).to(device)
self.src_mask = mask
else:
self.src_mask = None
src = self.encoder(src) * math.sqrt(self.ninp)
src = self.pos_encoder(src)
output = self.transformer_encoder(src, self.src_mask)
output = self.decoder(output)
return F.log_softmax(output, dim=-1)
| 0.936612 | 0.358185 |
Starting with `import` statements:
```
import pandas as pd
import psycopg2 as psy
from sqlalchemy import create_engine, inspect
from sqlalchemy.orm import sessionmaker
from sqlalchemy_utils import database_exists, create_database
from config import param_dic as param_dic
```
`param_dic` is a dictionary holding config info to connect to AWS DB
Creating a `connection` object using `psycopg2`:
```
conn = psy.connect(database=param_dic['db'], user=param_dic['user'],
host=param_dic['host'], password=param_dic['password'], port=param_dic['port'])
```
Creating a `cursor` **Factory**
```
def query_executer(query):
cursor_obj = conn.cursor()
cursor_obj.execute(query)
conn.commit()
```
Neccessary methods:
Creating a `session` *getter* method:
```
def get_session():
engine = get_engine(param_dic['user'], param_dic['password'], param_dic['host'], param_dic['port'], param_dic['db'])
session = sessionmaker(bind=engine)()
return session
```
Creating an `engine` *getter* method:
```
def get_engine(user, password, host, port, db):
engine_url = f"postgresql://{user}:{password}@{host}:{port}/{db}"
if not database_exists(engine_url):
create_database(engine_url)
sql_engine = create_engine(engine_url, pool_size=50, echo=False)
return sql_engine
```
Creating engine object:
```
engine = get_engine(param_dic['user'], param_dic['password'], param_dic['host'], param_dic['port'], param_dic['db'])
engine.connect()
session = get_session()
```
To list tables in a database, an inspector object is needed; as engine.table_names() is deprecated
```
inspector = inspect(engine)
print("Tables in Database:")
tables = inspector.get_table_names()
print(tables)
```
Creating Table **users** if it doesn't already exist:
```
if 'users' not in tables:
query_executer("CREATE TABLE users (id INT NOT NULL PRIMARY KEY,username VARCHAR(15) UNIQUE,fullname VARCHAR(50),email VARCHAR(100));")
print("TABLE CREATED")
else:
print("TABLE 'users' EXISTS !")
```
Storing **query** as pandas `DataFrame`:
```
query = "SELECT * FROM users"
```
Create a query object using `pandas.read_sql()` method:
```
query_obj = pd.read_sql(query, engine)
print(query_obj)
```
Inserting some data into the table with a function:
```
def insert_data(id, username, fullname, email):
try:
ins_query = f'INSERT INTO users VALUES({id}, \'{username}\', \'{fullname}\', \'{email}\')'
query_executer(ins_query)
except Exception as e:
print('Exception: ' + str(e))
conn.rollback()
```
Using the `insert_data` method to INSERT 3 records into TABLE 'users':
```
insert_data(1,"thrasher502","Mohammed Darras","some-email@domain")
insert_data(2,"Eddie","Mohammed Darras 2","some-different-email@domain")
query = "SELECT * FROM users"
query_obj = pd.read_sql(query, engine)
print(query_obj)
```
Dynamic `id` (last id+1):
```
if query_obj.empty:
last_id = 1
else:
last_id = (query_obj.iloc[-1])['id']
insert_data(last_id + 1,"dynamicID","using iloc","whatever@domain")
```
Printing Table after data insertion:
```
query = "SELECT * FROM users"
query_obj = pd.read_sql(query, engine)
print(query_obj)
```
Query with `WHERE`:
```
query = "SELECT * FROM users WHERE username = 'thrasher502'"
query_obj = pd.read_sql(query, engine)
print(query_obj)
```
|
github_jupyter
|
import pandas as pd
import psycopg2 as psy
from sqlalchemy import create_engine, inspect
from sqlalchemy.orm import sessionmaker
from sqlalchemy_utils import database_exists, create_database
from config import param_dic as param_dic
conn = psy.connect(database=param_dic['db'], user=param_dic['user'],
host=param_dic['host'], password=param_dic['password'], port=param_dic['port'])
def query_executer(query):
cursor_obj = conn.cursor()
cursor_obj.execute(query)
conn.commit()
def get_session():
engine = get_engine(param_dic['user'], param_dic['password'], param_dic['host'], param_dic['port'], param_dic['db'])
session = sessionmaker(bind=engine)()
return session
def get_engine(user, password, host, port, db):
engine_url = f"postgresql://{user}:{password}@{host}:{port}/{db}"
if not database_exists(engine_url):
create_database(engine_url)
sql_engine = create_engine(engine_url, pool_size=50, echo=False)
return sql_engine
engine = get_engine(param_dic['user'], param_dic['password'], param_dic['host'], param_dic['port'], param_dic['db'])
engine.connect()
session = get_session()
inspector = inspect(engine)
print("Tables in Database:")
tables = inspector.get_table_names()
print(tables)
if 'users' not in tables:
query_executer("CREATE TABLE users (id INT NOT NULL PRIMARY KEY,username VARCHAR(15) UNIQUE,fullname VARCHAR(50),email VARCHAR(100));")
print("TABLE CREATED")
else:
print("TABLE 'users' EXISTS !")
query = "SELECT * FROM users"
query_obj = pd.read_sql(query, engine)
print(query_obj)
def insert_data(id, username, fullname, email):
try:
ins_query = f'INSERT INTO users VALUES({id}, \'{username}\', \'{fullname}\', \'{email}\')'
query_executer(ins_query)
except Exception as e:
print('Exception: ' + str(e))
conn.rollback()
insert_data(1,"thrasher502","Mohammed Darras","some-email@domain")
insert_data(2,"Eddie","Mohammed Darras 2","some-different-email@domain")
query = "SELECT * FROM users"
query_obj = pd.read_sql(query, engine)
print(query_obj)
if query_obj.empty:
last_id = 1
else:
last_id = (query_obj.iloc[-1])['id']
insert_data(last_id + 1,"dynamicID","using iloc","whatever@domain")
query = "SELECT * FROM users"
query_obj = pd.read_sql(query, engine)
print(query_obj)
query = "SELECT * FROM users WHERE username = 'thrasher502'"
query_obj = pd.read_sql(query, engine)
print(query_obj)
| 0.214198 | 0.723956 |
# Software Carpentry
# Welcome to Binder
This is where will do all our Python, Shell and Git live coding.
## Jupyter Lab
Let's quickly familiarise ourselves with the enironment ...
- the overal environment (ie your entire browser tab) is called:
*Jupyter Lab*
it contains menus, tabs, toolbars and a file browser
- Jupyter Lab allows you to *launch* files and application into the *Work Area*. Right now you probably have two tabs in the *Work Area* - this document an another tab called *Launcher*.
## Juptyer Notebooks
- this document is document is called an:
*Interactive Python Notebook* (or Notebook for short)
Notebooks are a documents (files) made up of a sequence of *cells* that contain either code (python in our case) or documentation (text, or formatted text called *markdown*).
### Cells
The three types of Cells are:
- *Markdown* - formatted text like this cell (with *italics*, **bold** and tables etc ...)
- *Raw* - like the following cell, and
- *Code* - (in our case) python code
Cells can be modified by simply clicking inside them and typing.
See if you can change the cell below by replacing *boring* with something more exciting.
#### Executing Cells
## Both *markdown* and *code* cells can be excuted,
Executing a *markdown* causes the *formatted* version of the cell to be **displayed**. Excuting a *code* cell causes the code to run and any results are displayed below the cell.
Any cell can be executed by *pressing* the play icon at the top of the document while the cell is highlighted.
You can also press **CTL-ENTER** to excute the active cell.
Go ahead and make some more changes to the cells above and execute them - what happens when you execute a *Raw* cell ?
#### Adding a Removing Cells
You can use the `+` (plus icon) at the top of the docuement to add a new cell, and the cell type drop down the change the type.
You can also use the A key to add cell *above* the current cell and the B key to add *below* the current cell.
Now add a couple of cell of your own ...
## Hello this is a heading
*ital
#### Code Cells
Code cells allow us to write (in our case Python) and run our code and see the results right inside the notebook.
The next cell is a code cell that contains Python code to add 4 numbers.
Try executing the cell and if get the right result, try some more/different numbers
```
3 + 2 + 3 + 8
```
## Let's save our work so for and Push to our changes to GitHub
### Saving
By pressing the save icon on the document (or File -> Save Notebook) we can save our work to our Binder environment.
### But what about Version Control and Git (wasn't that in the Workshop syllabus)
Since our binder environment will dissappear when we are no longer using is, we need cause our new version to be saved some where permanent.
Luckly we have a GitHub respository already connected to our current environment - however there are couple steps required to make our GitHub repository match the copy of the respository inside our Binder environment.
#### Git likes to know how you are
... otherwise it keeps complaining it cannot track who made what commits (changes).
To tell Git who you are, we need to do the following:
- Launch a Terminal sesson (File -> New -> Terminal, or you can use the *Laucher* tab
- At the command prompt, type: `git-setup`
This operation only needs to be done once per binder session.
#### Add your changed files to git's list of files to track
- at the same terminal prompt type: `git add .`
#### Tell Git to commit (record) this state as a version
- at the same terminal prompt type: `git commit -m "changes made inside binder"`
at this point git has added an additional version of your files to your repository inside your curren Binder environment. However, your repository on GitHub remains unchanges (you might like to go check).
#### Tell Git to push the new commit (version) to GitHub
- again at the same prompt type: `git push`
once you supply the correct GitHub usename and password, all your changes will be pushed.
Go check out your respository on github.com ...
|
github_jupyter
|
3 + 2 + 3 + 8
| 0.074362 | 0.808786 |
# Lecture 7 Quiz
##Question 1
A student wants to write into a file called myfile, without deleting its existing content. Which one of the following functions should he or she use?
* f = open('myfile', 'w')
* f = open('myfile', 'a')
* f = open('myfile', 'r')
* f = open('myfile', 'w+b')
##Question 2
Which of the following statements are true?
A) When you open a file for reading, if the file does not exist, an error occurs.
B) When you open a file for writing, if the file does not exist, a new file is created.
C) When you open a file for reading, if the file does not exist, the program will open an empty file.
D) When you open a file for writing, if the file exists, the existing file is overwritten with the new file.
E) When you open a file for writing, if the file does not exist, an error occurs.
* A, and E
* A, B, and D only
* D only
* A and B only
```
f = open('b.py', 'r')
```
True statments:
A) When you open a file for reading, if the file does not exist, an error occurs.
B) When you open a file for writing, if the file does not exist, a new file is created.
D) When you open a file for writing, if the file exists, the existing file is overwritten with the new file.
##Question 3
Examine the following three functions that take as argument a file name and return the extension of that file. For instance, if the file name is 'myfile.tar.gz' the returned value of the function should be 'gz'. If the file name has no extention, i.e. when the file name is just 'myfile', the function should return an empty string.
```
def get_extension1(filename):
return(filename.split(".")[-1])
def get_extension2(filename):
import os.path
return(os.path.splitext(filename)[1])
def get_extension3(filename):
return filename[filename.rfind('.'):][1:]
```
Which of the these functions are doing exactly what they are supposed to do according to the description above?
* get_extension3 only
* get_extension1 only
* get_extension1 and get_extension2
* All of them.
```
def get_extension1(filename):
return(filename.split(".")[-1])
def get_extension2(filename):
import os.path
return(os.path.splitext(filename)[1])
def get_extension3(filename):
return filename[filename.rfind('.'):][1:]
print "get_extension1"
print "get_extension1('filename'):%s" % get_extension1('filename')
print "get_extension1('filename.py'):%s" % get_extension1('filename.py')
print
print "get_extension2"
print "get_extension2('filename'):%s" % get_extension2('filename')
print "get_extension2('filename.py'):%s" % get_extension2('filename.py')
print
print "get_extension3"
print "get_extension3('filename'):%s" % get_extension3('filename')
print "get_extension3('filename.py'):%s" % get_extension3('filename.py')
```
##Question 4
A student is running a Python program like this:
```>python mergefasta.py myfile1.fa myfile2.fa```
In the mergefasta.py program the following lines are present:
```
import sys
tocheck=sys.argv[1]
```
What is the value of the variable tocheck?
* 'myfile1.fa'
* mergefasta.py
* 'mergefasta.py'
* python
1. a.py
```
# -*- coding: utf-8 -*-
# !/usr/bin/python
import sys
tocheck=sys.argv[1]
print "sys.argv[0]:", sys.argv[0]
print "sys.argv[1]:", sys.argv[1]
print "sys.argv[2]:", sys.argv[2]
```
2. b.py
```
# -*- coding: utf-8 -*-
# !/usr/bin/python
print 'b'
```
3. c.py
```
# -*- coding: utf-8 -*-
# !/usr/bin/python
print 'c'
```
After I run `python a.py b.py c.py`, it shows below:
```
sys.argv[0]: a.py
sys.argv[1]: b.py
sys.argv[2]: c.py
```
##Question 5
A student launches the Python interpreter from his home directory. His home directory contains another directory called 'mydir'. What will happen when he write the following code at the Python prompt:
```
>>> import os
>>> filenames = os.listdir('mydir')
>>> f= open(filenames[0])
```
* An error will be produced stating that the file to be opened does not exist.
* A variable f representing a file object will be created, and the first file in the directory 'mydir' will be opened for reading in text mode.
* A variable f representing a file object will be created, and the first file in the directory 'mydir' will be opened.
* An error will be produced stating that filename is not subscriptable.
Because of current directory is not 'mydir', which just is a child directory, An error will be produced stating that the file to be opened does not exist.
|
github_jupyter
|
f = open('b.py', 'r')
def get_extension1(filename):
return(filename.split(".")[-1])
def get_extension2(filename):
import os.path
return(os.path.splitext(filename)[1])
def get_extension3(filename):
return filename[filename.rfind('.'):][1:]
def get_extension1(filename):
return(filename.split(".")[-1])
def get_extension2(filename):
import os.path
return(os.path.splitext(filename)[1])
def get_extension3(filename):
return filename[filename.rfind('.'):][1:]
print "get_extension1"
print "get_extension1('filename'):%s" % get_extension1('filename')
print "get_extension1('filename.py'):%s" % get_extension1('filename.py')
print
print "get_extension2"
print "get_extension2('filename'):%s" % get_extension2('filename')
print "get_extension2('filename.py'):%s" % get_extension2('filename.py')
print
print "get_extension3"
print "get_extension3('filename'):%s" % get_extension3('filename')
print "get_extension3('filename.py'):%s" % get_extension3('filename.py')
import sys
tocheck=sys.argv[1]
# -*- coding: utf-8 -*-
# !/usr/bin/python
import sys
tocheck=sys.argv[1]
print "sys.argv[0]:", sys.argv[0]
print "sys.argv[1]:", sys.argv[1]
print "sys.argv[2]:", sys.argv[2]
# -*- coding: utf-8 -*-
# !/usr/bin/python
print 'b'
# -*- coding: utf-8 -*-
# !/usr/bin/python
print 'c'
sys.argv[0]: a.py
sys.argv[1]: b.py
sys.argv[2]: c.py
>>> import os
>>> filenames = os.listdir('mydir')
>>> f= open(filenames[0])
| 0.140956 | 0.704224 |
```
# 패키지 import
import numpy as np
import json
from PIL import Image
import matplotlib.pyplot as plt
%matplotlib inline
import torch
import torchvision
from torchvision import models, transforms
# 파이토치 버전 확인
print("PyTorch Version: ", torch.__version__)
print("Torchvision Version: ", torchvision.__version__)
# VGG-16 모델의 인스턴스 생성
use_pretended = True # 학습된 파라미터 사용
net = models.vgg16(pretrained=use_pretended)
net.eval() # 추론 모드(평가 모드)로 설정
```
### 입력 영상의 전처리 클래스 작성
```
# 입력 화상의 전처리 클래스
class BaseTransform():
"""
화상 크기 변경 및 색상 표준화
Attributes
-----------
resize : int
크기 변경 전의 화상 크기
mean : (R, G, B)
std : (R, G, B)
"""
def __init__(self, resize, mean, std):
self.base_transform = transforms.Compose([
transforms.Resize(resize), # 짧은 변의 길이가 resize크기
transforms.CenterCrop(resize), # 화상 중앙을 resize x resize로 자름
transforms.ToTensor(), # 토치 텐서로 변환
transforms.Normalize(mean, std) # 색상 정보 표준화
])
# 인스턴스를 생성한 후 그 인스턴스명으로 실행하면 __call__()이 실행됨.
def __call__(self, img):
return self.base_transform(img)
# 화상 전처리 확인
# 화상 읽기
image_file_path = ('./data/goldenretriever-3724972_1920.jpg')
img = Image.open(image_file_path) # [높이][너비][색RGB]
plt.imshow(img)
plt.show()
# 화상 전처리 및 처리된 화상 표시
resize = 224
mean = (0.485, 0.456, 0.406)
std = (0.229, 0.224, 0.225)
transform = BaseTransform(resize, mean, std)
img_transformed = transform(img) # torch.Size([3, 224, 224])
print(img_transformed)
# (색상, 높이, 너비) -> (높이, 너비, 색상)
# 0-1로 값을 제한하여 표시
img_transformed = img_transformed.numpy().transpose([1, 2, 0])
img_transformed = np.clip(img_transformed, 0, 1)
plt.imshow(img_transformed)
plt.show()
```
### 출력 결과로 라벨을 예측하는 후처리 클래스 생성
```
# ILSVRC 라벨 정보를 읽어 사전형 변수 생성
ILSVRC_class_index = json.load(open('./data/imagenet_class_index.json', 'r'))
ILSVRC_class_index
# 출력 결과에서 라벨을 예측하는 후처리 클래스
class ILSVRCPredictor():
def __init__(self, class_index):
self.class_index = class_index
def predict_max(self, out):
maxid = np.argmax(out.detach().numpy())
predicted_label_name = self.class_index[str(maxid)][1]
return predicted_label_name
```
### 학습된 VGG모델로 화상 예측
```
ILSVRC_class_index = json.load(open('./data/imagenet_class_index.json', 'r'))
# 인스턴스 생성
predictor = ILSVRCPredictor(ILSVRC_class_index)
# 입력 화상 읽기
image_file_path = './data/computer.jpg'
img = Image.open(image_file_path) # [높이][너비][RGB]
plt.imshow(img)
plt.show()
# 전처리 후 배치 크기의 차원 추가
transform = BaseTransform(resize, mean, std)
img_transformed = transform(img) # torch.Size([3,224,224])
inputs = img_transformed.unsqueeze_(0) # torch.Size([1,3,224,224])
# 모델이 입력하고 모델 출력을 라벨로 변환
out = net(inputs)
predicted_label_name = predictor.predict_max(out)
print(predicted_label_name)
```
|
github_jupyter
|
# 패키지 import
import numpy as np
import json
from PIL import Image
import matplotlib.pyplot as plt
%matplotlib inline
import torch
import torchvision
from torchvision import models, transforms
# 파이토치 버전 확인
print("PyTorch Version: ", torch.__version__)
print("Torchvision Version: ", torchvision.__version__)
# VGG-16 모델의 인스턴스 생성
use_pretended = True # 학습된 파라미터 사용
net = models.vgg16(pretrained=use_pretended)
net.eval() # 추론 모드(평가 모드)로 설정
# 입력 화상의 전처리 클래스
class BaseTransform():
"""
화상 크기 변경 및 색상 표준화
Attributes
-----------
resize : int
크기 변경 전의 화상 크기
mean : (R, G, B)
std : (R, G, B)
"""
def __init__(self, resize, mean, std):
self.base_transform = transforms.Compose([
transforms.Resize(resize), # 짧은 변의 길이가 resize크기
transforms.CenterCrop(resize), # 화상 중앙을 resize x resize로 자름
transforms.ToTensor(), # 토치 텐서로 변환
transforms.Normalize(mean, std) # 색상 정보 표준화
])
# 인스턴스를 생성한 후 그 인스턴스명으로 실행하면 __call__()이 실행됨.
def __call__(self, img):
return self.base_transform(img)
# 화상 전처리 확인
# 화상 읽기
image_file_path = ('./data/goldenretriever-3724972_1920.jpg')
img = Image.open(image_file_path) # [높이][너비][색RGB]
plt.imshow(img)
plt.show()
# 화상 전처리 및 처리된 화상 표시
resize = 224
mean = (0.485, 0.456, 0.406)
std = (0.229, 0.224, 0.225)
transform = BaseTransform(resize, mean, std)
img_transformed = transform(img) # torch.Size([3, 224, 224])
print(img_transformed)
# (색상, 높이, 너비) -> (높이, 너비, 색상)
# 0-1로 값을 제한하여 표시
img_transformed = img_transformed.numpy().transpose([1, 2, 0])
img_transformed = np.clip(img_transformed, 0, 1)
plt.imshow(img_transformed)
plt.show()
# ILSVRC 라벨 정보를 읽어 사전형 변수 생성
ILSVRC_class_index = json.load(open('./data/imagenet_class_index.json', 'r'))
ILSVRC_class_index
# 출력 결과에서 라벨을 예측하는 후처리 클래스
class ILSVRCPredictor():
def __init__(self, class_index):
self.class_index = class_index
def predict_max(self, out):
maxid = np.argmax(out.detach().numpy())
predicted_label_name = self.class_index[str(maxid)][1]
return predicted_label_name
ILSVRC_class_index = json.load(open('./data/imagenet_class_index.json', 'r'))
# 인스턴스 생성
predictor = ILSVRCPredictor(ILSVRC_class_index)
# 입력 화상 읽기
image_file_path = './data/computer.jpg'
img = Image.open(image_file_path) # [높이][너비][RGB]
plt.imshow(img)
plt.show()
# 전처리 후 배치 크기의 차원 추가
transform = BaseTransform(resize, mean, std)
img_transformed = transform(img) # torch.Size([3,224,224])
inputs = img_transformed.unsqueeze_(0) # torch.Size([1,3,224,224])
# 모델이 입력하고 모델 출력을 라벨로 변환
out = net(inputs)
predicted_label_name = predictor.predict_max(out)
print(predicted_label_name)
| 0.584864 | 0.924959 |
```
%matplotlib inline
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import accuracy_score
import numpy as np
import scipy.io as io
import matplotlib.pyplot as plt
plt.style.use('fivethirtyeight')
plt.rcParams['font.family'] = 'sans-serif'
plt.rcParams['font.serif'] = 'Ubuntu'
plt.rcParams['font.monospace'] = 'Ubuntu Mono'
plt.rcParams['font.size'] = 10
plt.rcParams['axes.labelsize'] = 10
plt.rcParams['axes.labelweight'] = 'bold'
plt.rcParams['axes.titlesize'] = 10
plt.rcParams['xtick.labelsize'] = 8
plt.rcParams['ytick.labelsize'] = 8
plt.rcParams['legend.fontsize'] = 10
plt.rcParams['figure.titlesize'] = 12
plt.rcParams['image.cmap'] = 'jet'
plt.rcParams['image.interpolation'] = 'none'
plt.rcParams['figure.figsize'] = (16, 8)
plt.rcParams['lines.linewidth'] = 2
colors = ['#008fd5', '#fc4f30', '#e5ae38', '#6d904f', '#8b8b8b', '#810f7c', '#137e6d', '#be0119', '#3b638c', '#af6f09',
'#c79fef', '#80f9ad']
```
Legge il dataset
```
data = io.loadmat("XwindowsDocData.mat")
xtrain = data['xtrain']
ytrain = data['ytrain']
ndocs = xtrain.shape[0]
nterms = xtrain.shape[1]
classes = np.unique(ytrain)
nclasses = classes.shape[0]
```
Crea un classificatore Naive-Bayes ed effettua il learning sul dataset
```
clf = MultinomialNB(alpha=0)
clf.fit(xtrain, ytrain.ravel())
```
Deriva probabilità a priori delle classi $p(C_i)$
```
pclass=np.exp(clf.class_log_prior_)
```
Deriva probabilità a posteriori delle feature (termini) $p(t_j|C_i)$
```
pf=np.exp(clf.feature_log_prob_)
```
Deriva probabilità a priori delle feature $p(t_j)=p(t_j|C_1)p(C_1)+p(t_j|C_2)p(C_2)$
```
pfeature=np.array([pf[0,i]*pclass[0]+pf[1,i]*pclass[1] for i in range(pf.shape[1])])
```
Plot del modello di linguaggio risultante per le due classi
```
fig=plt.figure(figsize=(16,8))
for i,c in enumerate(classes):
ax=plt.subplot(nclasses,1,i+1)
ax.bar(range(clf.feature_count_.shape[1]), pf[i,:], facecolor=colors[i], alpha=0.9, edgecolor=colors[i], lw=2)
plt.title('$p(t_j|C_{0:1d})$'.format(c))
plt.suptitle('Language models by ML, no smoothing ')
plt.show()
```
Applicazione del classificatore al training set e derivazione della accuracy
```
preds = clf.predict(xtrain)
print('Accuracy = {0:8.7f}'.format(accuracy_score(ytrain, preds)))
```
Istanza classificatore Naive-Bayes con Dirichlet smoothing simmetrico, con iperparametro $\alpha$ ed effettua il learning sul dataset
```
alpha = 10
clf1 = MultinomialNB(alpha)
clf1.fit(xtrain, ytrain.ravel())
```
Deriva probabilità a posteriori delle feature (termini) $p(t_j|C_i)$
```
pf1=np.exp(clf1.feature_log_prob_)
```
Plot del modello di linguaggio risultante per le due classi
```
fig=plt.figure(figsize=(16,8))
for i,c in enumerate(classes):
ax=plt.subplot(nclasses,1,i+1)
ax.bar(range(clf1.feature_count_.shape[1]), pf1[i,:], facecolor=colors[i+2], alpha=0.9, edgecolor=colors[i+2], lw=2)
plt.title('$p(t_j|C_{0:1d})$'.format(c))
plt.suptitle(r"Language models by bayesian learning, uniform dirichlet, $\alpha= {0:2d}$".format(alpha))
plt.show()
```
Applicazione del classificatore al training set e derivazione della accuracy
```
preds1 = clf1.predict(xtrain)
print('Accuracy = {0:8.7f}'.format(accuracy_score(ytrain, preds1)))
```
Definizione della funzione che calcola la mutua informazione di una feature con le due classi
```
def mutual_information(feature):
s = 0
for cl in [0,1]:
s += pf[cl, feature]*pclass[cl]*np.log2(pf[cl, feature]/pfeature[feature])
return s
```
Calcolo del valore della mutua informazione per ogni feature
```
mi = np.array([mutual_information(f) for f in range(pf.shape[1])])
```
Ordinamento crescente delle feature rispetto alla mutua informazione
```
ordered_features = np.argsort(mi)
```
Plot della mutua informazione delle feature
```
fig=plt.figure(figsize=(16,8))
plt.bar(range(clf1.feature_count_.shape[1]), mi, facecolor=colors[5], alpha=0.9, edgecolor=colors[5], lw=2)
plt.title(r"Mutual information")
plt.show()
```
Seleziona le feature più informative e riduci il training set considerando solo quelle
```
k = 100
x_red = xtrain[:, ordered_features[-k:]]
```
Plot della mutua informazione delle feature selezionate
```
fig=plt.figure(figsize=(16,8))
plt.bar(range(k), mi[ordered_features[-k:]], facecolor=colors[6], alpha=0.9, edgecolor=colors[6], lw=2)
plt.title(r"Mutual information")
plt.show()
```
Crea un nuovo classificatore Naive Bayes ed effettua l'apprendimento sul training set ridotto
```
clf2 = MultinomialNB(alpha=0)
clf2.fit(x_red, ytrain.ravel())
```
Applicazione del classificatore al training set ridotto e derivazione della accuracy
```
preds2 = clf2.predict(x_red)
print('Accuracy = {0:8.7f}'.format(accuracy_score(ytrain, preds2)))
```
|
github_jupyter
|
%matplotlib inline
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import accuracy_score
import numpy as np
import scipy.io as io
import matplotlib.pyplot as plt
plt.style.use('fivethirtyeight')
plt.rcParams['font.family'] = 'sans-serif'
plt.rcParams['font.serif'] = 'Ubuntu'
plt.rcParams['font.monospace'] = 'Ubuntu Mono'
plt.rcParams['font.size'] = 10
plt.rcParams['axes.labelsize'] = 10
plt.rcParams['axes.labelweight'] = 'bold'
plt.rcParams['axes.titlesize'] = 10
plt.rcParams['xtick.labelsize'] = 8
plt.rcParams['ytick.labelsize'] = 8
plt.rcParams['legend.fontsize'] = 10
plt.rcParams['figure.titlesize'] = 12
plt.rcParams['image.cmap'] = 'jet'
plt.rcParams['image.interpolation'] = 'none'
plt.rcParams['figure.figsize'] = (16, 8)
plt.rcParams['lines.linewidth'] = 2
colors = ['#008fd5', '#fc4f30', '#e5ae38', '#6d904f', '#8b8b8b', '#810f7c', '#137e6d', '#be0119', '#3b638c', '#af6f09',
'#c79fef', '#80f9ad']
data = io.loadmat("XwindowsDocData.mat")
xtrain = data['xtrain']
ytrain = data['ytrain']
ndocs = xtrain.shape[0]
nterms = xtrain.shape[1]
classes = np.unique(ytrain)
nclasses = classes.shape[0]
clf = MultinomialNB(alpha=0)
clf.fit(xtrain, ytrain.ravel())
pclass=np.exp(clf.class_log_prior_)
pf=np.exp(clf.feature_log_prob_)
pfeature=np.array([pf[0,i]*pclass[0]+pf[1,i]*pclass[1] for i in range(pf.shape[1])])
fig=plt.figure(figsize=(16,8))
for i,c in enumerate(classes):
ax=plt.subplot(nclasses,1,i+1)
ax.bar(range(clf.feature_count_.shape[1]), pf[i,:], facecolor=colors[i], alpha=0.9, edgecolor=colors[i], lw=2)
plt.title('$p(t_j|C_{0:1d})$'.format(c))
plt.suptitle('Language models by ML, no smoothing ')
plt.show()
preds = clf.predict(xtrain)
print('Accuracy = {0:8.7f}'.format(accuracy_score(ytrain, preds)))
alpha = 10
clf1 = MultinomialNB(alpha)
clf1.fit(xtrain, ytrain.ravel())
pf1=np.exp(clf1.feature_log_prob_)
fig=plt.figure(figsize=(16,8))
for i,c in enumerate(classes):
ax=plt.subplot(nclasses,1,i+1)
ax.bar(range(clf1.feature_count_.shape[1]), pf1[i,:], facecolor=colors[i+2], alpha=0.9, edgecolor=colors[i+2], lw=2)
plt.title('$p(t_j|C_{0:1d})$'.format(c))
plt.suptitle(r"Language models by bayesian learning, uniform dirichlet, $\alpha= {0:2d}$".format(alpha))
plt.show()
preds1 = clf1.predict(xtrain)
print('Accuracy = {0:8.7f}'.format(accuracy_score(ytrain, preds1)))
def mutual_information(feature):
s = 0
for cl in [0,1]:
s += pf[cl, feature]*pclass[cl]*np.log2(pf[cl, feature]/pfeature[feature])
return s
mi = np.array([mutual_information(f) for f in range(pf.shape[1])])
ordered_features = np.argsort(mi)
fig=plt.figure(figsize=(16,8))
plt.bar(range(clf1.feature_count_.shape[1]), mi, facecolor=colors[5], alpha=0.9, edgecolor=colors[5], lw=2)
plt.title(r"Mutual information")
plt.show()
k = 100
x_red = xtrain[:, ordered_features[-k:]]
fig=plt.figure(figsize=(16,8))
plt.bar(range(k), mi[ordered_features[-k:]], facecolor=colors[6], alpha=0.9, edgecolor=colors[6], lw=2)
plt.title(r"Mutual information")
plt.show()
clf2 = MultinomialNB(alpha=0)
clf2.fit(x_red, ytrain.ravel())
preds2 = clf2.predict(x_red)
print('Accuracy = {0:8.7f}'.format(accuracy_score(ytrain, preds2)))
| 0.488771 | 0.873053 |
# Group 50: Neighboorhoods Similar to Thneedville
# Introduction
Love-the-life-we-lead-ville? Destined-to-succeed-ville? We-are-all-agreed-ville? You guessed it! Our overall metric for our final project is to find the neighborhood that best mirrors or reflects Thneedville from the Lorax. The inspiration behind choosing the datasets we chose were based on the story and the elements of the book/movie and lyrics of the song on the Original Soundtrack that shares the same name, Thneedville. Water quality, budgeting for capital projects, and tree benefits were assessed to analyze which neighborhood was the most similar to Thneedville from The Lorax.
# Elevated Blood Lead Levels in Allegheny County
The first metric we'll be looking at elevated blood lead levels in children in Allegheny County. In the song that shares the same name as the town, Thneedville, one part of the song references a little boy that now glows after swimming. This metric assesses water quality in various neighborhoods in or around Pittsburgh.

```
import pandas as pd
import numpy as np
import geopandas as gdp
%matplotlib inline
import matplotlib.pyplot as plt
blood_lead_levels = pd.read_csv("wprdc_ebll.csv")
lead_levels = gdp.read_file("EBLL_CT15_19.shp")
pd.options.display.max_columns = None
lead_levels.plot()
blood_lead_levels.columns
```
**Census Tract Blood Lead Level Test Results Visualized**
<br>The shp file contains geographical data, which is useful to then use geopandas to graph. This is the initial graph generated using geopandas and the shp file for the chosen dataset. The designated zones given by the geoid's in the spreadsheet. Geoids are number codes given by the Census Bureau. Some geoids may correspond to various art
located within the same neighborhood, so there may be a bit of overlap in the data.
There is a column that contains the average elevated blood lead levels from 2015 to 2020, so the columns that have notes for each years are removed to just include the data.
```
data = blood_lead_levels.drop(['note2020', 'note2019','note2018','note2017','note2016','note2015', 'note15_20'], axis = 1 )
```
The columns that give a note for the EBLL values for each years is removed.
```
updated_values = data.sort_values(by = 'percentEBLL15_20', ascending = False)
important_info = updated_values.head(25)
important_info
```
Since the resulting data gives geoid numbers, they have to be converted to the actual name of the neighborhood, which can be looked up.
42003561200 = Wilkinsburg, PA <br>
42003550900 = McKeesport, PA <br>
42003300100 = Knocksville, PA <br>
42003552300 = McKeesport, PA <br>
42003120700 = Homewood West <br>
42003261400 = Perry South <br>
42003552000 = McKeesport <br>
42003261500 = Perrysville, PA/Ross Township <br>
42003561000 = Wilkinsburg, PA <br>
42003512800 = North Braddock <br>
42003271500 = Marshall-Shadeland(Woods Run) <br>
42003130200 = Homewood North <br>
42003270300 = Brighton Heights <br>
42003241200 = Spring Garden <br>
42003250300 = Mexican War Streets/Northside <br>
42003010300 = Bluff(Uptown) <br>
42003260700 = Perry North <br>
42003120400 = Larimer <br>
42003562500 = Esplen/Sheriden <br>
42003111400 = Garfield <br>
42003561700 = St. Clair <br>
42003180300 = Allentown <br>
42003130400 = Homewood South <br>
42003562400 = Beltzhoover/Bon Air <br>
42003508000 = Wilmerding <br>
After ordering the values from greatest to least, the neighborhoods were converted from geoid to a physical location.
```
wilkinsburg_average = (15.79 + 11.04)/2
mckeesport_average = (14.77 + 12.99 + 11.76)/3
print(wilkinsburg_average)
print(mckeesport_average)
```
Since Wilkinsburg and McKeesport appear multiple times in the data, the average is calculated for the final graph.
```
final_ebll = pd.DataFrame({'Neighborhoods': ['Wilkinsburg','McKeesport', 'Knocksville', 'Homewood West', 'Perry South',
'Perrysville/Ross Township', 'North Braddock', 'Marshall-Shadeland(Woods Run)',
'Homwood North', 'Brighton Heights', 'Spring Garden', 'Mexican War Streets',
'Bluff(Uptown)', 'Perry North', 'Larimer', 'Esplen/Sheriden','Garfield','St. Clair'
, 'Allentown', 'Homewood South', 'Belzhoover/Bon Air', 'Wilmerding'],
'percentEBLL15_20': [13.42,13.17, 14.17, 12.20, 11.82, 11.28,10.84,10.45,10.37,
10.29, 10.29,10.26, 10.20,10.06,9.84, 9.62, 9.52, 9.26,
9.25, 9.15, 8.85, 8.70]})
final_ebll.plot.bar(figsize = (10,10), x = 'Neighborhoods', y = 'percentEBLL15_20')
```
A new data frame containing the average elevated blood lead levels is created.
**Analysis and Summary of Data**
<br> Overall, the neighborhoods in Pittsburgh and surrounding area that have the highest blood lead levels are either in lower income areas or are in North Side Pittsburgh. Multiple Census Tract numbers that are traceable to Wilkinsburg and McKeesport have a larger recorded percentage of blood lead levels. However, the neighborhood that has the overall highest blood lead level in Knocksville, which has a average percentage of blood lead levels from 2015 to 2020. Although the water quality isn't quite the same as Thneedville, an increased concentration in lead in water can vause adverse effects, especially in children.
# Capital Projects
```
import pandas as pd
import geopandas
```
The town in the Lorax, Thneedville, was set up entirely as a result of capital greed. To find the neighborhood that best matches Thneedville, I will look at the captial projects throughout Pittsburgh, and find the best.
```
capital = pd.read_csv("capitalprojects.csv", sep = ',', index_col = 'id')
capital.sample(5)
capital.groupby(['neighborhood']).mean()
capital.describe()
```
On average, the neighborhoods in Pittsburgh are spending around *1.94 million* dollars on each capital project.
## Neighborhoods with Highest Budgets
To get a better sense of which neighborhoods are spending the most, we'll look at the total spending of each neighborhood, as well as the average they're spending on each project.
```
capital_projects = capital.groupby("neighborhood").sum()['budgeted_amount']
budget_capital = capital_projects.sort_values(ascending = False)
budget_capital.head()
projects_neighborhood = capital.groupby("neighborhood").mean()['budgeted_amount']
projects_neighborhood.sort_values(ascending = False).head()
```
Unlike the total budget of each neighborhood, *Knoxville*, tops the chart at around **1.45 million dollars** per project. Greenfield, however, has the highest total budget for captial projects.
There are a lot of NaN, so we'll drop the columns that don't mean much to our metric.
```
capital_df = capital.dropna()
capital_df.sample(5)
#mask to only look at neighborhoods above threshold
project_mask = capital_df['budgeted_amount'] >= 1200000
high_budget = capital_df[project_mask]
high_budget.shape[0]
high_budget.sort_values(by = 'budgeted_amount', ascending = False).head(7)
#create mask to look at individual neighborhoods
greenfield_mask = capital_df['neighborhood'] == 'Greenfield'
greenfield_df = capital_df[greenfield_mask]
greenfield_df['status'].value_counts()
greenfield_df['status'].value_counts().sort_index().plot.pie()
south_mask = capital_df['neighborhood'] == 'South Side Flats'
south_df = capital_df[south_mask]
south_df['status'].value_counts()
south_df['status'].value_counts().sort_index().plot.pie()
#mask to keep in progress and completed
complete_project = capital_df['status'] != 'Planned'
complete_df = capital_df[complete_project]
total_budget = complete_df.groupby('neighborhood').sum()['budgeted_amount']
neighborhoods = geopandas.read_file("Neighborhoods_.shp")
#plot based on total budget for each neighborhood
capital_map = neighborhoods.merge(total_budget, how='left', left_on='hood', right_on='neighborhood')
capital_map.dropna()
capital_map[['hood', 'budgeted_amount', 'geometry']]
capital_map.plot(column = 'budgeted_amount',
cmap = 'OrRd',
edgecolor = 'black',
legend = True,
legend_kwds = {'label': 'Budget'})
total_budget.sort_values(ascending = False)
sum_budget = total_budget.sum()
total_budget.div(sum_budget).mul(100).sort_values(ascending = False)
```
My way of normalizing the data was to make each a percentage. This allowed for us to combine our metrics into an equally weighted category.
The highest budget for all capital projects in a neighborhood is in **Squirrel Hill North** with a budget of around **4 Million dollars**. South Side Flats and Central Business District follow with budgets of 2.8 million and 2.6 million respectively.
Given this information, the best neighborhood in Pittsburgh is **Squirrel Hill North**. It best resembles Thneedville from the Lorax, given it's high capital budget.
# Tree Impact in Neighborhoods
This metric looks at trees managed by the city of Pittsburgh itself. Neighorhoods may have more trees than listed, but they aren't owned by the city, and therefore can be bought and cut down to build the neighborhood's capitalism. None of those trees are safe from capitalism; We're only looking at the ones that are (for now). The neighborhood with the fewest city-protected tree benefits will most resemble Thneedville.
```
import pandas as pd
trees = pd.read_csv("https://data.wprdc.org/datastore/dump/1515a93c-73e3-4425-9b35-1cd11b2196da")
hoods = pd.read_csv("https://data.wprdc.org/datastore/dump/668d7238-cfd2-492e-b397-51a6e74182ff")
trees2cols = pd.DataFrame({'airtotalval' : trees.groupby( 'neighborhood' )['air_quality_benfits_total_lbs'].agg('sum')}).reset_index()
trees2cols.head(3)
```
We are only looking at the air quality benefits in pounds per neighborhood. The above code gets rid of superfluous columns. Here's a graph of this data, where more yellow areas have more pounds of benefits:
```
import geopandas
pittsburghmap = geopandas.read_file("https://data.wprdc.org/dataset/e672f13d-71c4-4a66-8f38-710e75ed80a4/resource/c5a93a8e-03d7-4eb3-91a8-c6b7db0fa261/download/dbd133a206cc4a3aa915cb28baa60fd4_0.zip")
airmap = pittsburghmap.merge(trees2cols, how = 'left', left_on = 'hood', right_on = 'neighborhood')
airmap.plot(column = "airtotalval", cmap = "summer", legend = True, figsize = (15,15), legend_kwds = {'label': "Pounds of Air Quality Benefits"})
```
To standardize this data, we can take the area in acres of each neighborhood and divide the neighborhood's total pounds of air quality benefits by its size to get a better representation. The following code also combines the dataframe with the neighborhood sizes with the pounds of air benefits value into a final data frame.
```
hoods2cols = pd.DataFrame( {'areaacres' : hoods.groupby('hood')['acres'].agg('sum')}).reset_index()
finalframe = pd.merge(trees2cols, hoods2cols, left_on="neighborhood", right_on="hood" ).drop("hood", axis=1)
finalframe.head(3)
```
The below code adds a missing value to the table and does the mentioned calculations to find the pounds of benefits per acre for each neighborhood. The data is sorted from least pounds per acre to greatest, since we're looking for neighborhoods with the LEAST amount of benefits.
```
finalframe.at[60, "areaacres"] = 775.68
finalframe["poundsperacre"] = finalframe["airtotalval"].div(finalframe["areaacres"].values)
finalframe.sort_values(by = 'poundsperacre', ascending = True).head(10)
```
To come up with a final score to combine with the scores of other metrics, I used the percentile for each data point. Each neighborhood has a score between 0-100. This helped make the close-together data more distinguishable. The scores with higher values have more pounds per acre.
```
finalframe['treescore'] = 100 - (pd.qcut(finalframe['poundsperacre'], q = 10000, labels = False))/100
finalframe.sort_values(by = 'treescore', ascending = False).head(25)
```
The above neighborhoods are the top twenty-five neighborhoods for the metric of least pounds of air benefits per acre in a neighborhood. Below is this information plotted on a map, where the more yellow areas have the highest calculated score:
```
import geopandas
pittsburghmap = geopandas.read_file("https://data.wprdc.org/dataset/e672f13d-71c4-4a66-8f38-710e75ed80a4/resource/c5a93a8e-03d7-4eb3-91a8-c6b7db0fa261/download/dbd133a206cc4a3aa915cb28baa60fd4_0.zip")
scoremap = pittsburghmap.merge(finalframe, how = 'left', left_on = 'hood', right_on = 'neighborhood')
scoremap.plot(column = "treescore", cmap = "summer", legend = True, figsize = (15,15), legend_kwds = {'label': "Calculated Score"})
```
##### Conclusion
The large, dark-green areas to the east contain many of the city's parks, so they have the highest density of air quality benefits. The areas with the least air quality benefits from government owned trees are ones further from the center of the city, because these areas have plenty of privately-owned trees to be consumed by capitalism in the future. Therefore, any neighborhood in Pittsburgh that is not near the center of the city and doesn't have vast public parks most resembles Thneedville, and has the lowest density of air-improving trees that are protected by the government.
# Combined Metric
Since three different datasets were used to pinpoint and select one distinct neighborhood that best reflects Thneedville, the combined metric will be using data points from those neighborhoods in the last metric, since trees are the most important aspect of the book and the movie. The top 25 neighborhoods with the highest treescore will be used in this final metric. Since all three metrics have percentages calculated, the neighborhood with the highest percentage will be the best neighborhood. If any of our other metrics do not include a certain neighborhood in our top values, the value for that specific metric will be a zero.
```
final_metric = pd.DataFrame({'Neighborhood': ['Hays', 'Glen Hazel', 'South Shore', 'Esplen', 'East Carnegie', 'Northview Heights'
, 'St. Clair', 'Arlington Heights', 'Fairywood', 'Ridgemont', 'Mt. Oliver', 'Arlington',
'Duquesne Heights', 'Troy Hill', 'Spring Garden', 'Spring Hill', 'West Oakland', 'South Side Slopes',
'Summer Hill', 'Allentown','South Oakland', 'Beltzhoover', 'Central Lawrenceville', 'Bluff', 'Bon Air'],
'PercentEBLL15_20': [0, 0, 0, 9.62, 0, 0, 9.26, 0, 0, 0, 0, 0, 0, 0, 10.29, 0, 0, 0, 0, 9.25, 0, 8.85, 0, 10.20, 8.85],
'Total_Budget':[1.712940, 0, 0, 0.139776, 0, 0, 0, 0, 0.017815, 0, 0, 0, 0, 0.502166, 0, 1.490258, 0, 0, 2.534124, 0.428235,
0.420099, 4.047421, 0.102776, 0, 0],
'Treescore': [100.00, 98.88, 97.76, 96.63, 95.51, 94.39, 93.26, 92.14, 91.02, 89.89, 88.77, 87.65, 86.52, 85.40, 84.27,
83.15, 82.03, 80.90, 79.78, 78.66, 77.53, 76.41, 75.29, 74.16, 73.04]})
final_metric.plot.bar(stacked=True, title='Metric Total', color=("black", "yellow", "green"), x = 'Neighborhood', figsize = (10,10))
```
According to the graph, Esplen is the best neighborhood, followed by St. Clair, Spring Garden, Beltzhoover, and Allentown.
## Conclusion/Summary
Overall, when combining all of the metrics, Esplen, according to our tests, is the most similar to Thneedville from The Lorax. All three metrics didn't consistently have the same neighborhoods, so the neighborhoods listed for the highest treescore was used because the significance of trees in The Lorax was prioritized. The most contributing factor in calculating the metric total was the treesore, since the average elevated blood lead levels and capital budgeting percentage were significantly lower. However, with those two values considered, some neighborhoods didn't have a score for either metric. Reflecting on this project, because of the metrics we measured we weren't going to have the same neighborhoods collectively, since a metric like elevated blood lead levels primarily had lower income neighborhoods, versus a metric like capital budgeting for projects, which includes affluent areas where a significant amount of money can be allocated to those projects.
|
github_jupyter
|
import pandas as pd
import numpy as np
import geopandas as gdp
%matplotlib inline
import matplotlib.pyplot as plt
blood_lead_levels = pd.read_csv("wprdc_ebll.csv")
lead_levels = gdp.read_file("EBLL_CT15_19.shp")
pd.options.display.max_columns = None
lead_levels.plot()
blood_lead_levels.columns
data = blood_lead_levels.drop(['note2020', 'note2019','note2018','note2017','note2016','note2015', 'note15_20'], axis = 1 )
updated_values = data.sort_values(by = 'percentEBLL15_20', ascending = False)
important_info = updated_values.head(25)
important_info
wilkinsburg_average = (15.79 + 11.04)/2
mckeesport_average = (14.77 + 12.99 + 11.76)/3
print(wilkinsburg_average)
print(mckeesport_average)
final_ebll = pd.DataFrame({'Neighborhoods': ['Wilkinsburg','McKeesport', 'Knocksville', 'Homewood West', 'Perry South',
'Perrysville/Ross Township', 'North Braddock', 'Marshall-Shadeland(Woods Run)',
'Homwood North', 'Brighton Heights', 'Spring Garden', 'Mexican War Streets',
'Bluff(Uptown)', 'Perry North', 'Larimer', 'Esplen/Sheriden','Garfield','St. Clair'
, 'Allentown', 'Homewood South', 'Belzhoover/Bon Air', 'Wilmerding'],
'percentEBLL15_20': [13.42,13.17, 14.17, 12.20, 11.82, 11.28,10.84,10.45,10.37,
10.29, 10.29,10.26, 10.20,10.06,9.84, 9.62, 9.52, 9.26,
9.25, 9.15, 8.85, 8.70]})
final_ebll.plot.bar(figsize = (10,10), x = 'Neighborhoods', y = 'percentEBLL15_20')
import pandas as pd
import geopandas
capital = pd.read_csv("capitalprojects.csv", sep = ',', index_col = 'id')
capital.sample(5)
capital.groupby(['neighborhood']).mean()
capital.describe()
capital_projects = capital.groupby("neighborhood").sum()['budgeted_amount']
budget_capital = capital_projects.sort_values(ascending = False)
budget_capital.head()
projects_neighborhood = capital.groupby("neighborhood").mean()['budgeted_amount']
projects_neighborhood.sort_values(ascending = False).head()
capital_df = capital.dropna()
capital_df.sample(5)
#mask to only look at neighborhoods above threshold
project_mask = capital_df['budgeted_amount'] >= 1200000
high_budget = capital_df[project_mask]
high_budget.shape[0]
high_budget.sort_values(by = 'budgeted_amount', ascending = False).head(7)
#create mask to look at individual neighborhoods
greenfield_mask = capital_df['neighborhood'] == 'Greenfield'
greenfield_df = capital_df[greenfield_mask]
greenfield_df['status'].value_counts()
greenfield_df['status'].value_counts().sort_index().plot.pie()
south_mask = capital_df['neighborhood'] == 'South Side Flats'
south_df = capital_df[south_mask]
south_df['status'].value_counts()
south_df['status'].value_counts().sort_index().plot.pie()
#mask to keep in progress and completed
complete_project = capital_df['status'] != 'Planned'
complete_df = capital_df[complete_project]
total_budget = complete_df.groupby('neighborhood').sum()['budgeted_amount']
neighborhoods = geopandas.read_file("Neighborhoods_.shp")
#plot based on total budget for each neighborhood
capital_map = neighborhoods.merge(total_budget, how='left', left_on='hood', right_on='neighborhood')
capital_map.dropna()
capital_map[['hood', 'budgeted_amount', 'geometry']]
capital_map.plot(column = 'budgeted_amount',
cmap = 'OrRd',
edgecolor = 'black',
legend = True,
legend_kwds = {'label': 'Budget'})
total_budget.sort_values(ascending = False)
sum_budget = total_budget.sum()
total_budget.div(sum_budget).mul(100).sort_values(ascending = False)
import pandas as pd
trees = pd.read_csv("https://data.wprdc.org/datastore/dump/1515a93c-73e3-4425-9b35-1cd11b2196da")
hoods = pd.read_csv("https://data.wprdc.org/datastore/dump/668d7238-cfd2-492e-b397-51a6e74182ff")
trees2cols = pd.DataFrame({'airtotalval' : trees.groupby( 'neighborhood' )['air_quality_benfits_total_lbs'].agg('sum')}).reset_index()
trees2cols.head(3)
import geopandas
pittsburghmap = geopandas.read_file("https://data.wprdc.org/dataset/e672f13d-71c4-4a66-8f38-710e75ed80a4/resource/c5a93a8e-03d7-4eb3-91a8-c6b7db0fa261/download/dbd133a206cc4a3aa915cb28baa60fd4_0.zip")
airmap = pittsburghmap.merge(trees2cols, how = 'left', left_on = 'hood', right_on = 'neighborhood')
airmap.plot(column = "airtotalval", cmap = "summer", legend = True, figsize = (15,15), legend_kwds = {'label': "Pounds of Air Quality Benefits"})
hoods2cols = pd.DataFrame( {'areaacres' : hoods.groupby('hood')['acres'].agg('sum')}).reset_index()
finalframe = pd.merge(trees2cols, hoods2cols, left_on="neighborhood", right_on="hood" ).drop("hood", axis=1)
finalframe.head(3)
finalframe.at[60, "areaacres"] = 775.68
finalframe["poundsperacre"] = finalframe["airtotalval"].div(finalframe["areaacres"].values)
finalframe.sort_values(by = 'poundsperacre', ascending = True).head(10)
finalframe['treescore'] = 100 - (pd.qcut(finalframe['poundsperacre'], q = 10000, labels = False))/100
finalframe.sort_values(by = 'treescore', ascending = False).head(25)
import geopandas
pittsburghmap = geopandas.read_file("https://data.wprdc.org/dataset/e672f13d-71c4-4a66-8f38-710e75ed80a4/resource/c5a93a8e-03d7-4eb3-91a8-c6b7db0fa261/download/dbd133a206cc4a3aa915cb28baa60fd4_0.zip")
scoremap = pittsburghmap.merge(finalframe, how = 'left', left_on = 'hood', right_on = 'neighborhood')
scoremap.plot(column = "treescore", cmap = "summer", legend = True, figsize = (15,15), legend_kwds = {'label': "Calculated Score"})
final_metric = pd.DataFrame({'Neighborhood': ['Hays', 'Glen Hazel', 'South Shore', 'Esplen', 'East Carnegie', 'Northview Heights'
, 'St. Clair', 'Arlington Heights', 'Fairywood', 'Ridgemont', 'Mt. Oliver', 'Arlington',
'Duquesne Heights', 'Troy Hill', 'Spring Garden', 'Spring Hill', 'West Oakland', 'South Side Slopes',
'Summer Hill', 'Allentown','South Oakland', 'Beltzhoover', 'Central Lawrenceville', 'Bluff', 'Bon Air'],
'PercentEBLL15_20': [0, 0, 0, 9.62, 0, 0, 9.26, 0, 0, 0, 0, 0, 0, 0, 10.29, 0, 0, 0, 0, 9.25, 0, 8.85, 0, 10.20, 8.85],
'Total_Budget':[1.712940, 0, 0, 0.139776, 0, 0, 0, 0, 0.017815, 0, 0, 0, 0, 0.502166, 0, 1.490258, 0, 0, 2.534124, 0.428235,
0.420099, 4.047421, 0.102776, 0, 0],
'Treescore': [100.00, 98.88, 97.76, 96.63, 95.51, 94.39, 93.26, 92.14, 91.02, 89.89, 88.77, 87.65, 86.52, 85.40, 84.27,
83.15, 82.03, 80.90, 79.78, 78.66, 77.53, 76.41, 75.29, 74.16, 73.04]})
final_metric.plot.bar(stacked=True, title='Metric Total', color=("black", "yellow", "green"), x = 'Neighborhood', figsize = (10,10))
| 0.297368 | 0.902093 |
# Retrieve and prepare data
```
#nbd module
import io
import numpy as np
import pandas as pd
import geopandas
from popemp.tools import Nbd, download_file
nbd = Nbd('popemp')
data_dir = nbd.root/'data'
```
# Geography
[geocodes](https://www.census.gov/geographies/reference-files/2019/demo/popest/2019-fips.html)
```
#nbd module
def geo():
df_file = data_dir/'geo.pkl'
if df_file.exists():
return pd.read_pickle(df_file)
f = download_file('https://www2.census.gov/geo/tiger/GENZ2018/shp/cb_2018_us_state_20m.zip', data_dir)
df = geopandas.read_file(f)
df = df.rename(columns={'STATEFP': 'st', 'NAME': 'name'})
df = df[['st', 'name', 'geometry']]
df['cty'] = '000'
st = df
f = download_file('https://www2.census.gov/geo/tiger/GENZ2018/shp/cb_2018_us_county_20m.zip', data_dir)
df = geopandas.read_file(f)
df = df.rename(columns={'STATEFP': 'st', 'COUNTYFP': 'cty', 'NAME': 'name_cty'})
df = df[['st', 'cty', 'name_cty', 'geometry']]
df = df.merge(st[['st', 'name']], 'left')
df['name'] = df['name_cty'] + ' county, ' + df['name']
del df['name_cty']
df = pd.concat([df, st]).sort_values(['st', 'cty'], ignore_index=True)
df = df[['st', 'cty', 'name', 'geometry']]
df.to_pickle(df_file)
return df
d = geo()
d.head()
```
# Population
[home](https://www.census.gov/programs-surveys/popest/data/data-sets.html)
[2000-2010](https://www.census.gov/data/datasets/time-series/demo/popest/intercensal-2000-2010-counties.html)
[2010-2019](https://www.census.gov/data/datasets/time-series/demo/popest/2010s-counties-total.html)
For years before 1990 I could not find data in convenient format.
[Character encoding](https://www.census.gov/programs-surveys/geography/technical-documentation/user-note/special-characters.html): newer files use "UTF-8", older use "ISO-8859-1".
```
#nbd module
def pop_1990_1999():
f = download_file('https://www2.census.gov/programs-surveys/popest/tables/1990-2000/estimates-and-change-1990-2000/2000c8_00.txt', data_dir)
with open(f, encoding='ISO-8859-1') as file:
data = io.StringIO()
in_table = False
for line in file:
if in_table:
if line[0] == '1':
data.write(line)
else:
break
else:
if line[0] == '1':
in_table = True
data.write(line)
data.seek(0)
df = pd.read_fwf(data, dtype='str', header=None)
# skip first row (US total), keep fips and popest cols
df = df.iloc[1:, 1:13]
df.columns = ['fips'] + [f'pop{y}' for y in range(2000, 1989, -1)]
df['fips'] = df['fips'].str.pad(5, 'right', '0')
df['st'] = df['fips'].str[:2]
df['cty'] = df['fips'].str[2:]
df = df.drop(columns=['pop2000', 'fips'])
df = pd.wide_to_long(df, 'pop', ['st', 'cty'], 'year')
df = df.reset_index()
df['pop'] = pd.to_numeric(df['pop'].str.replace(',', '', regex=False)).astype('int')
return df
#nbd module
def pop_2000_2009():
f = download_file('https://www2.census.gov/programs-surveys/popest/datasets/2000-2010/intercensal/county/co-est00int-tot.csv', data_dir)
cols = ['STATE', 'COUNTY'] + [f'POPESTIMATE{y}' for y in range(2000, 2010)]
df = pd.read_csv(f, encoding='ISO-8859-1', dtype='str', usecols=cols)
df = pd.wide_to_long(df, 'POPESTIMATE', ['STATE', 'COUNTY'], 'year')
df = df.reset_index().rename(columns={'STATE': 'st', 'COUNTY': 'cty', 'POPESTIMATE': 'pop'})
df['st'] = df['st'].str.pad(2, fillchar='0')
df['cty'] = df['cty'].str.pad(3, fillchar='0')
df['pop'] = df['pop'].astype('int')
return df
#nbd module
def pop_2010_2019():
f = download_file('https://www2.census.gov/programs-surveys/popest/datasets/2010-2019/counties/totals/co-est2019-alldata.csv', data_dir)
cols = ['STATE', 'COUNTY'] + [f'POPESTIMATE{y}' for y in range(2010, 2020)]
df = pd.read_csv(f, encoding='ISO-8859-1', dtype='str', usecols=cols)
df = pd.wide_to_long(df, 'POPESTIMATE', ['STATE', 'COUNTY'], 'year')
df = df.reset_index().rename(columns={'STATE': 'st', 'COUNTY': 'cty', 'POPESTIMATE': 'pop'})
df['pop'] = df['pop'].astype('int')
return df
#nbd module
def pop():
df_file = data_dir/'pop.pkl'
if df_file.exists():
return pd.read_pickle(df_file)
d1 = pop_1990_1999()
d2 = pop_2000_2009()
d3 = pop_2010_2019()
df = pd.concat([d1, d2, d3], ignore_index=True)
d = df.query('cty == "000"').groupby('year')['pop'].sum()
d = d.to_frame('pop').reset_index()
d[['st', 'cty']] = ['00', '000']
df = pd.concat([df, d], ignore_index=True)
df = df.sort_values('year')
df['pop_'] = df.groupby(['st', 'cty'])['pop'].shift()
df['pop_gr'] = df.eval('(pop / pop_ - 1) * 100')
del df['pop_']
df = df.sort_values(['st', 'cty', 'year'])
df = df[['st', 'cty', 'year', 'pop', 'pop_gr']]
df.to_pickle(df_file)
return df
```
Something strange happens in 2000.
```
d = pop()
d.query('st == "00" and cty == "000"').set_index('year')['pop'].plot()
d.query('st == "01" and cty == "000"').set_index('year')['pop'].plot()
```
# Employment
[datasets](https://www.census.gov/data/datasets/time-series/econ/bds/bds-datasets.html)
```
#nbd module
def emp():
df_file = data_dir/'emp.pkl'
if df_file.exists():
return pd.read_pickle(df_file)
# economy-wide
f = download_file('https://www2.census.gov/programs-surveys/bds/tables/time-series/bds2019.csv', data_dir)
df = pd.read_csv(f, usecols=['year', 'emp', 'net_job_creation_rate'], dtype='str')
df[['st', 'cty']] = ['00', '000']
d1 = df
# by state
f = download_file('https://www2.census.gov/programs-surveys/bds/tables/time-series/bds2019_st.csv', data_dir)
df = pd.read_csv(f, usecols=['year', 'st', 'emp', 'net_job_creation_rate'], dtype='str')
df['cty'] = '000'
d2 = df
# by county
f = download_file('https://www2.census.gov/programs-surveys/bds/tables/time-series/bds2019_cty.csv', data_dir)
df = pd.read_csv(f, usecols=['year', 'st', 'cty', 'emp', 'net_job_creation_rate'], dtype='str')
df = pd.concat([d1, d2, df], ignore_index=True)
df = df.rename(columns={'net_job_creation_rate': 'emp_gr'})
df['year'] = df['year'].astype('int16')
df['emp'] = pd.to_numeric(df['emp'], 'coerce')
df['emp_gr'] = pd.to_numeric(df['emp_gr'], 'coerce')
df = df[['st', 'cty', 'year', 'emp', 'emp_gr']]
df.to_pickle(df_file)
return df
```
## API
[BDS API](https://www.census.gov/data/developers/data-sets/business-dynamics.html)
```
key = open('census_api_key.txt').read()
url = 'https://api.census.gov/data/timeseries/bds'
st = '55'
r = requests.get(f'{url}?get=NAME,ESTAB,EMP,YEAR&for=county:*&in=state:{st}&time=from+2015+to+2019&NAICS=00&key={key}')
d = r.json()
df = pd.DataFrame(d[1:], columns=d[0])
df.query('county == "025"')
```
# Build this module
```
nbd.nb2mod('data.ipynb')
```
|
github_jupyter
|
#nbd module
import io
import numpy as np
import pandas as pd
import geopandas
from popemp.tools import Nbd, download_file
nbd = Nbd('popemp')
data_dir = nbd.root/'data'
#nbd module
def geo():
df_file = data_dir/'geo.pkl'
if df_file.exists():
return pd.read_pickle(df_file)
f = download_file('https://www2.census.gov/geo/tiger/GENZ2018/shp/cb_2018_us_state_20m.zip', data_dir)
df = geopandas.read_file(f)
df = df.rename(columns={'STATEFP': 'st', 'NAME': 'name'})
df = df[['st', 'name', 'geometry']]
df['cty'] = '000'
st = df
f = download_file('https://www2.census.gov/geo/tiger/GENZ2018/shp/cb_2018_us_county_20m.zip', data_dir)
df = geopandas.read_file(f)
df = df.rename(columns={'STATEFP': 'st', 'COUNTYFP': 'cty', 'NAME': 'name_cty'})
df = df[['st', 'cty', 'name_cty', 'geometry']]
df = df.merge(st[['st', 'name']], 'left')
df['name'] = df['name_cty'] + ' county, ' + df['name']
del df['name_cty']
df = pd.concat([df, st]).sort_values(['st', 'cty'], ignore_index=True)
df = df[['st', 'cty', 'name', 'geometry']]
df.to_pickle(df_file)
return df
d = geo()
d.head()
#nbd module
def pop_1990_1999():
f = download_file('https://www2.census.gov/programs-surveys/popest/tables/1990-2000/estimates-and-change-1990-2000/2000c8_00.txt', data_dir)
with open(f, encoding='ISO-8859-1') as file:
data = io.StringIO()
in_table = False
for line in file:
if in_table:
if line[0] == '1':
data.write(line)
else:
break
else:
if line[0] == '1':
in_table = True
data.write(line)
data.seek(0)
df = pd.read_fwf(data, dtype='str', header=None)
# skip first row (US total), keep fips and popest cols
df = df.iloc[1:, 1:13]
df.columns = ['fips'] + [f'pop{y}' for y in range(2000, 1989, -1)]
df['fips'] = df['fips'].str.pad(5, 'right', '0')
df['st'] = df['fips'].str[:2]
df['cty'] = df['fips'].str[2:]
df = df.drop(columns=['pop2000', 'fips'])
df = pd.wide_to_long(df, 'pop', ['st', 'cty'], 'year')
df = df.reset_index()
df['pop'] = pd.to_numeric(df['pop'].str.replace(',', '', regex=False)).astype('int')
return df
#nbd module
def pop_2000_2009():
f = download_file('https://www2.census.gov/programs-surveys/popest/datasets/2000-2010/intercensal/county/co-est00int-tot.csv', data_dir)
cols = ['STATE', 'COUNTY'] + [f'POPESTIMATE{y}' for y in range(2000, 2010)]
df = pd.read_csv(f, encoding='ISO-8859-1', dtype='str', usecols=cols)
df = pd.wide_to_long(df, 'POPESTIMATE', ['STATE', 'COUNTY'], 'year')
df = df.reset_index().rename(columns={'STATE': 'st', 'COUNTY': 'cty', 'POPESTIMATE': 'pop'})
df['st'] = df['st'].str.pad(2, fillchar='0')
df['cty'] = df['cty'].str.pad(3, fillchar='0')
df['pop'] = df['pop'].astype('int')
return df
#nbd module
def pop_2010_2019():
f = download_file('https://www2.census.gov/programs-surveys/popest/datasets/2010-2019/counties/totals/co-est2019-alldata.csv', data_dir)
cols = ['STATE', 'COUNTY'] + [f'POPESTIMATE{y}' for y in range(2010, 2020)]
df = pd.read_csv(f, encoding='ISO-8859-1', dtype='str', usecols=cols)
df = pd.wide_to_long(df, 'POPESTIMATE', ['STATE', 'COUNTY'], 'year')
df = df.reset_index().rename(columns={'STATE': 'st', 'COUNTY': 'cty', 'POPESTIMATE': 'pop'})
df['pop'] = df['pop'].astype('int')
return df
#nbd module
def pop():
df_file = data_dir/'pop.pkl'
if df_file.exists():
return pd.read_pickle(df_file)
d1 = pop_1990_1999()
d2 = pop_2000_2009()
d3 = pop_2010_2019()
df = pd.concat([d1, d2, d3], ignore_index=True)
d = df.query('cty == "000"').groupby('year')['pop'].sum()
d = d.to_frame('pop').reset_index()
d[['st', 'cty']] = ['00', '000']
df = pd.concat([df, d], ignore_index=True)
df = df.sort_values('year')
df['pop_'] = df.groupby(['st', 'cty'])['pop'].shift()
df['pop_gr'] = df.eval('(pop / pop_ - 1) * 100')
del df['pop_']
df = df.sort_values(['st', 'cty', 'year'])
df = df[['st', 'cty', 'year', 'pop', 'pop_gr']]
df.to_pickle(df_file)
return df
d = pop()
d.query('st == "00" and cty == "000"').set_index('year')['pop'].plot()
d.query('st == "01" and cty == "000"').set_index('year')['pop'].plot()
#nbd module
def emp():
df_file = data_dir/'emp.pkl'
if df_file.exists():
return pd.read_pickle(df_file)
# economy-wide
f = download_file('https://www2.census.gov/programs-surveys/bds/tables/time-series/bds2019.csv', data_dir)
df = pd.read_csv(f, usecols=['year', 'emp', 'net_job_creation_rate'], dtype='str')
df[['st', 'cty']] = ['00', '000']
d1 = df
# by state
f = download_file('https://www2.census.gov/programs-surveys/bds/tables/time-series/bds2019_st.csv', data_dir)
df = pd.read_csv(f, usecols=['year', 'st', 'emp', 'net_job_creation_rate'], dtype='str')
df['cty'] = '000'
d2 = df
# by county
f = download_file('https://www2.census.gov/programs-surveys/bds/tables/time-series/bds2019_cty.csv', data_dir)
df = pd.read_csv(f, usecols=['year', 'st', 'cty', 'emp', 'net_job_creation_rate'], dtype='str')
df = pd.concat([d1, d2, df], ignore_index=True)
df = df.rename(columns={'net_job_creation_rate': 'emp_gr'})
df['year'] = df['year'].astype('int16')
df['emp'] = pd.to_numeric(df['emp'], 'coerce')
df['emp_gr'] = pd.to_numeric(df['emp_gr'], 'coerce')
df = df[['st', 'cty', 'year', 'emp', 'emp_gr']]
df.to_pickle(df_file)
return df
key = open('census_api_key.txt').read()
url = 'https://api.census.gov/data/timeseries/bds'
st = '55'
r = requests.get(f'{url}?get=NAME,ESTAB,EMP,YEAR&for=county:*&in=state:{st}&time=from+2015+to+2019&NAICS=00&key={key}')
d = r.json()
df = pd.DataFrame(d[1:], columns=d[0])
df.query('county == "025"')
nbd.nb2mod('data.ipynb')
| 0.268941 | 0.801081 |
```
1
2
1+2
1.0
1.
1.*2
1/2
1//2
2-3
2**3
import math
help(math)
(1+2)*3
dir(math)
dir(1.0)
math.log2(8)
'star'
'shooting'+'star'
'star'*3
'star'[3]
'star'[0]
'star'[0:4]
'star'[:2]
'star'[1:]
'star'[-1]
'star'[:-2]
y = 'star'
y
y[0]
n = -1
y[n]
y = 3
z = 10
y+z
y, z = 3, 10
y
z
my_list = [1, 2, 3]
my_list2 = ['a', 1, 'b', 'train', 3.0]
my_list+my_list2
my_list*3
my_list[2]
len(my_list)
n = len(my_list)
n
math.log2(8)
import numpy as np
my_array = np.array(my_list)
my_other_array = np.array([1, 3, 4])
my_list
my_array
my_list + my_list
my_array + my_array
type(1.)
type(1)
type(my_list)
type(my_array)
```
### Exercise
1. Creat your own numpy array using only numbers.
2. Experiment with different arithmetic options.
3. How do arrays behave differently from lists.
4. Does the numpy module have a mean funciton? How do you know?
5. Find the mean of your array.
```
my_array = np.array([1, 2, 3, 4])
print(my_array*3)
print(my_array**2)
my_list = [1, 3, 'ddd', 1.3]
my_array = np.array([1, '54', 'hello', 32.0])
my_array
my_array = np.array([1, 2, 3, 4])
np.mean(my_array)
np.mean(my_array)
my_array.mean()
my_array = np.array([[1, 2, 3],
[2, 2, 4]])
my_array
my_array.mean()
my_array.mean(axis=1)
my_array
my_array.size
my_array.shape
dir(my_array)
help(my_array)
np.argmax?
my_array = np.array([[1, 2, 3],
[1, 2, 4]])
my_array
my_array.mean(axis=0)
my_array[0]
my_array[0,0]
my_array[0:2, 0]
```
# Here's some math
$$ \int_0^x x^2 dx $$
the transpose of [my array is](www.google.com):
```
my_array.T
```
# Heading
### Smaller heading
[links](http://google.com)
* lists
* items
* tada
```
my_array
my_array.shape
my_favorite_stars = ['sirius', 'deneb', 'altair']
for star in my_favorite_stars:
print('One of my favorite stars is {}'.format(star))
constellation = 'lyra'
if constellation == 'cygnus':
print('cygnus is my favorite constellation')
else:
print('{} is not my favorite constellation'.format(constellation))
'{} is {} is {} is {}'.format(1, 2, 3, 4)
```
## Exercise
Loop through your list of favorite stars. If the name has 6 letters, print the name of the star. Otherwise, print a message that the number of letters in the star name was the wrong size.
```
for star in my_favorite_stars:
if len(star) == 6:
print(star)
else:
print('Length of name {} is too short'.format(star))
len(star) == 6
print(star)
True
False
if 0:
print('True or False')
if None:
print('This is 2')
my_list = [1, 2, 5, 7, 9, 4, 5, 4]
# Find the total
total = 0
for number in my_list:
total = total + number
# calculate the mean
mean = total/len(my_list)
# Find the median
sorted_list = sorted(my_list)
if len(my_list)%2==1:
median_index = int(len(my_list)/2)
median = sorted_list[median_index]
else:
median_index = int(len(my_list)/2)
median = (sorted_list[median_index]+sorted_list[median_index-1])/2
print(mean, median)
print(np.mean(my_list), np.median(my_list))
3//2
int(1.5)
def calculate_total(my_list):
"""Calculate the total of a list.
Parameters
----------
my_list : list
This is the list to calculate a total.
Returns
-------
total : int/float
This is the total of our list.
"""
total = 0
for number in my_list:
total += number
return total
def calculate_mean(my_list):
total = calculate_total(my_list)
list_length = len(my_list)
mean = total/list_length
return mean
def calculate_median(my_list):
sorted_list = sorted(my_list)
if len(my_list)%2==1:
median_index = int(len(my_list)/2)
median = sorted_list[median_index]
else:
median_index = int(len(my_list)/2)
median = (sorted_list[median_index]+ sorted_list[median_index-1])/2
return median
my_list = [1, 2, 5, 7, 9, 4, 5, 4]
mean = calculate_mean(my_list)
median = calculate_median(my_list)
print(mean, median)
help(calculate_total)
```
## Exercise
Write a function that takes a distance in parsecs and converts it to a distance in megaparsecs.
```
def convert_to_megaparsecs(distance_pc):
"""Convert the given distance from parsecs
to megaparsecs.
Parameters
----------
distance_pc : int,float
The distance in parsecs.
Returns
-------
distance_mpc : int, float
The distance in megaparsecs.
"""
distance_mpc = distance_pc/10**6
return distance_mpc
distance = 30000000
convert_to_megaparsecs(distance)
data_dir = 'path_to_your_repo/2019-01-05-aas/python'
data_file = 'hubble_data.dat'
from astropy.table import Table
from astropy.units import u
import os
full_data_file = os.path.join(data_dir, data_file)
full_data_file
import glob
glob.glob(full_data_file)
from astropy.table import Table
import astropy.units as u
table_data = Table.read(full_data_file,
names = ['galaxy',
'supernova',
'm', 'sig_m',
'dist_mod',
'sig_dist_mod',
'M', 'sig_M',
'velocity'],
format='ascii'
)
import astropy.table
table_data
table_data[0]
table_data['supernova']
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
%matplotlib inline
plt.plot(table_data['dist_mod'],
table_data['velocity'], '^', color='green')
```
## Exercise:
Write a function to convert distance modulus into distance.
That equation is ...
$$DistMod = m - M = 5*log(d) - 5$$
```
def find_distance(dist_mod):
"""Find a distance given a distance modulus.
Parameters
----------
dist_mod : int,float
Distance modulus.
Returns
-------
distance : int, float
The distance in parsecs.
"""
distance = 10**((dist_mod+5)/5)*u.pc
return distance
dist_pc = find_distance(table_data['dist_mod'])
dist_pc
help(convert_to_megaparsecs)
dist_mpc = convert_to_megaparsecs(dist_pc)
dist_mpc
def convert_to_megaparsecs2(distance):
dist_mpc = distance.to(u.Mpc)
return dist_mpc
dist_pc = find_distance(table_data['dist_mod'])*u.pc
dist_mpc = convert_to_megaparsecs2(dist_pc)
dist_mpc
plt.plot(dist_mpc, table_data['velocity'], 'x')
plt.plot(dist_mpc, np.polyval(fit_velocity, dist_mpc))
fit_velocity = np.polyfit(dist_mpc,
table_data['velocity'],
1)
print(fit_velocity)
# Plot data with label
plt.plot(dist_mpc, table_data['velocity'],
'x', label='Modulus Data')
plt.plot(dist_mpc,
np.polyval(fit_velocity, dist_mpc),
label='Fit')
# Label axes
plt.xlabel('Distance (Mpc)')
plt.ylabel(r'Velcity ($\frac{km}{s}$)')
# Add legend
plt.legend(loc=2)
# Title
plt.title('Hubble Diagram')
# Save
plt.savefig('hubble_diagram.png')
```
|
github_jupyter
|
1
2
1+2
1.0
1.
1.*2
1/2
1//2
2-3
2**3
import math
help(math)
(1+2)*3
dir(math)
dir(1.0)
math.log2(8)
'star'
'shooting'+'star'
'star'*3
'star'[3]
'star'[0]
'star'[0:4]
'star'[:2]
'star'[1:]
'star'[-1]
'star'[:-2]
y = 'star'
y
y[0]
n = -1
y[n]
y = 3
z = 10
y+z
y, z = 3, 10
y
z
my_list = [1, 2, 3]
my_list2 = ['a', 1, 'b', 'train', 3.0]
my_list+my_list2
my_list*3
my_list[2]
len(my_list)
n = len(my_list)
n
math.log2(8)
import numpy as np
my_array = np.array(my_list)
my_other_array = np.array([1, 3, 4])
my_list
my_array
my_list + my_list
my_array + my_array
type(1.)
type(1)
type(my_list)
type(my_array)
my_array = np.array([1, 2, 3, 4])
print(my_array*3)
print(my_array**2)
my_list = [1, 3, 'ddd', 1.3]
my_array = np.array([1, '54', 'hello', 32.0])
my_array
my_array = np.array([1, 2, 3, 4])
np.mean(my_array)
np.mean(my_array)
my_array.mean()
my_array = np.array([[1, 2, 3],
[2, 2, 4]])
my_array
my_array.mean()
my_array.mean(axis=1)
my_array
my_array.size
my_array.shape
dir(my_array)
help(my_array)
np.argmax?
my_array = np.array([[1, 2, 3],
[1, 2, 4]])
my_array
my_array.mean(axis=0)
my_array[0]
my_array[0,0]
my_array[0:2, 0]
my_array.T
my_array
my_array.shape
my_favorite_stars = ['sirius', 'deneb', 'altair']
for star in my_favorite_stars:
print('One of my favorite stars is {}'.format(star))
constellation = 'lyra'
if constellation == 'cygnus':
print('cygnus is my favorite constellation')
else:
print('{} is not my favorite constellation'.format(constellation))
'{} is {} is {} is {}'.format(1, 2, 3, 4)
for star in my_favorite_stars:
if len(star) == 6:
print(star)
else:
print('Length of name {} is too short'.format(star))
len(star) == 6
print(star)
True
False
if 0:
print('True or False')
if None:
print('This is 2')
my_list = [1, 2, 5, 7, 9, 4, 5, 4]
# Find the total
total = 0
for number in my_list:
total = total + number
# calculate the mean
mean = total/len(my_list)
# Find the median
sorted_list = sorted(my_list)
if len(my_list)%2==1:
median_index = int(len(my_list)/2)
median = sorted_list[median_index]
else:
median_index = int(len(my_list)/2)
median = (sorted_list[median_index]+sorted_list[median_index-1])/2
print(mean, median)
print(np.mean(my_list), np.median(my_list))
3//2
int(1.5)
def calculate_total(my_list):
"""Calculate the total of a list.
Parameters
----------
my_list : list
This is the list to calculate a total.
Returns
-------
total : int/float
This is the total of our list.
"""
total = 0
for number in my_list:
total += number
return total
def calculate_mean(my_list):
total = calculate_total(my_list)
list_length = len(my_list)
mean = total/list_length
return mean
def calculate_median(my_list):
sorted_list = sorted(my_list)
if len(my_list)%2==1:
median_index = int(len(my_list)/2)
median = sorted_list[median_index]
else:
median_index = int(len(my_list)/2)
median = (sorted_list[median_index]+ sorted_list[median_index-1])/2
return median
my_list = [1, 2, 5, 7, 9, 4, 5, 4]
mean = calculate_mean(my_list)
median = calculate_median(my_list)
print(mean, median)
help(calculate_total)
def convert_to_megaparsecs(distance_pc):
"""Convert the given distance from parsecs
to megaparsecs.
Parameters
----------
distance_pc : int,float
The distance in parsecs.
Returns
-------
distance_mpc : int, float
The distance in megaparsecs.
"""
distance_mpc = distance_pc/10**6
return distance_mpc
distance = 30000000
convert_to_megaparsecs(distance)
data_dir = 'path_to_your_repo/2019-01-05-aas/python'
data_file = 'hubble_data.dat'
from astropy.table import Table
from astropy.units import u
import os
full_data_file = os.path.join(data_dir, data_file)
full_data_file
import glob
glob.glob(full_data_file)
from astropy.table import Table
import astropy.units as u
table_data = Table.read(full_data_file,
names = ['galaxy',
'supernova',
'm', 'sig_m',
'dist_mod',
'sig_dist_mod',
'M', 'sig_M',
'velocity'],
format='ascii'
)
import astropy.table
table_data
table_data[0]
table_data['supernova']
from matplotlib import pyplot as plt
import matplotlib.pyplot as plt
%matplotlib inline
plt.plot(table_data['dist_mod'],
table_data['velocity'], '^', color='green')
def find_distance(dist_mod):
"""Find a distance given a distance modulus.
Parameters
----------
dist_mod : int,float
Distance modulus.
Returns
-------
distance : int, float
The distance in parsecs.
"""
distance = 10**((dist_mod+5)/5)*u.pc
return distance
dist_pc = find_distance(table_data['dist_mod'])
dist_pc
help(convert_to_megaparsecs)
dist_mpc = convert_to_megaparsecs(dist_pc)
dist_mpc
def convert_to_megaparsecs2(distance):
dist_mpc = distance.to(u.Mpc)
return dist_mpc
dist_pc = find_distance(table_data['dist_mod'])*u.pc
dist_mpc = convert_to_megaparsecs2(dist_pc)
dist_mpc
plt.plot(dist_mpc, table_data['velocity'], 'x')
plt.plot(dist_mpc, np.polyval(fit_velocity, dist_mpc))
fit_velocity = np.polyfit(dist_mpc,
table_data['velocity'],
1)
print(fit_velocity)
# Plot data with label
plt.plot(dist_mpc, table_data['velocity'],
'x', label='Modulus Data')
plt.plot(dist_mpc,
np.polyval(fit_velocity, dist_mpc),
label='Fit')
# Label axes
plt.xlabel('Distance (Mpc)')
plt.ylabel(r'Velcity ($\frac{km}{s}$)')
# Add legend
plt.legend(loc=2)
# Title
plt.title('Hubble Diagram')
# Save
plt.savefig('hubble_diagram.png')
| 0.528047 | 0.778965 |
## Dependencies
```
!pip install --quiet /kaggle/input/kerasapplications
!pip install --quiet /kaggle/input/efficientnet-git
import warnings, glob
from tensorflow.keras import Sequential, Model
import efficientnet.tfkeras as efn
from cassava_scripts import *
seed = 0
seed_everything(seed)
warnings.filterwarnings('ignore')
```
### Hardware configuration
```
# TPU or GPU detection
# Detect hardware, return appropriate distribution strategy
strategy, tpu = set_up_strategy()
AUTO = tf.data.experimental.AUTOTUNE
REPLICAS = strategy.num_replicas_in_sync
print(f'REPLICAS: {REPLICAS}')
```
# Model parameters
```
BATCH_SIZE = 8 * REPLICAS
HEIGHT = 512
WIDTH = 512
CHANNELS = 3
N_CLASSES = 5
TTA_STEPS = 0 # Do TTA if > 0
```
# Augmentation
```
def data_augment(image, label):
p_spatial = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_rotate = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_1 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_2 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_3 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_crop = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
# Flips
image = tf.image.random_flip_left_right(image)
image = tf.image.random_flip_up_down(image)
if p_spatial > .75:
image = tf.image.transpose(image)
# Rotates
if p_rotate > .75:
image = tf.image.rot90(image, k=3) # rotate 270º
elif p_rotate > .5:
image = tf.image.rot90(image, k=2) # rotate 180º
elif p_rotate > .25:
image = tf.image.rot90(image, k=1) # rotate 90º
# Pixel-level transforms
if p_pixel_1 >= .4:
image = tf.image.random_saturation(image, lower=.7, upper=1.3)
if p_pixel_2 >= .4:
image = tf.image.random_contrast(image, lower=.8, upper=1.2)
if p_pixel_3 >= .4:
image = tf.image.random_brightness(image, max_delta=.1)
# Crops
if p_crop > .7:
if p_crop > .9:
image = tf.image.central_crop(image, central_fraction=.7)
elif p_crop > .8:
image = tf.image.central_crop(image, central_fraction=.8)
else:
image = tf.image.central_crop(image, central_fraction=.9)
elif p_crop > .4:
crop_size = tf.random.uniform([], int(HEIGHT*.8), HEIGHT, dtype=tf.int32)
image = tf.image.random_crop(image, size=[crop_size, crop_size, CHANNELS])
# # Crops
# if p_crop > .6:
# if p_crop > .9:
# image = tf.image.central_crop(image, central_fraction=.5)
# elif p_crop > .8:
# image = tf.image.central_crop(image, central_fraction=.6)
# elif p_crop > .7:
# image = tf.image.central_crop(image, central_fraction=.7)
# else:
# image = tf.image.central_crop(image, central_fraction=.8)
# elif p_crop > .3:
# crop_size = tf.random.uniform([], int(HEIGHT*.6), HEIGHT, dtype=tf.int32)
# image = tf.image.random_crop(image, size=[crop_size, crop_size, CHANNELS])
return image, label
```
## Auxiliary functions
```
# Datasets utility functions
def resize_image(image, label):
image = tf.image.resize(image, [HEIGHT, WIDTH])
image = tf.reshape(image, [HEIGHT, WIDTH, CHANNELS])
return image, label
def process_path(file_path):
name = get_name(file_path)
img = tf.io.read_file(file_path)
img = decode_image(img)
img, _ = scale_image(img, None)
# img = center_crop(img, HEIGHT, WIDTH)
return img, name
def get_dataset(files_path, shuffled=False, tta=False, extension='jpg'):
dataset = tf.data.Dataset.list_files(f'{files_path}*{extension}', shuffle=shuffled)
dataset = dataset.map(process_path, num_parallel_calls=AUTO)
if tta:
dataset = dataset.map(data_augment, num_parallel_calls=AUTO)
dataset = dataset.map(resize_image, num_parallel_calls=AUTO)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.prefetch(AUTO)
return dataset
```
# Load data
```
database_base_path = '/kaggle/input/cassava-leaf-disease-classification/'
submission = pd.read_csv(f'{database_base_path}sample_submission.csv')
display(submission.head())
TEST_FILENAMES = tf.io.gfile.glob(f'{database_base_path}test_tfrecords/ld_test*.tfrec')
NUM_TEST_IMAGES = count_data_items(TEST_FILENAMES)
print(f'GCS: test: {NUM_TEST_IMAGES}')
model_path_list = glob.glob('/kaggle/input/44-cassava-leaf-effnetb5-2020-sing-cut-out-512x512/*.h5')
model_path_list.sort()
print('Models to predict:')
print(*model_path_list, sep='\n')
```
# Model
```
def model_fn(input_shape, N_CLASSES):
inputs = L.Input(shape=input_shape, name='input_image')
base_model = efn.EfficientNetB5(input_tensor=inputs,
include_top=False,
weights=None,
pooling='avg')
x = L.Dropout(.25)(base_model.output)
output = L.Dense(N_CLASSES, activation='softmax', name='output')(x)
model = Model(inputs=inputs, outputs=output)
return model
with strategy.scope():
model = model_fn((None, None, CHANNELS), N_CLASSES)
model.summary()
```
# Test set predictions
```
files_path = f'{database_base_path}test_images/'
test_size = len(os.listdir(files_path))
test_preds = np.zeros((test_size, N_CLASSES))
for model_path in model_path_list:
print(model_path)
K.clear_session()
model.load_weights(model_path)
if TTA_STEPS > 0:
test_ds = get_dataset(files_path, tta=True).repeat()
ct_steps = TTA_STEPS * ((test_size/BATCH_SIZE) + 1)
preds = model.predict(test_ds, steps=ct_steps, verbose=1)[:(test_size * TTA_STEPS)]
preds = np.mean(preds.reshape(test_size, TTA_STEPS, N_CLASSES, order='F'), axis=1)
test_preds += preds / len(model_path_list)
else:
test_ds = get_dataset(files_path, tta=False)
x_test = test_ds.map(lambda image, image_name: image)
test_preds += model.predict(x_test) / len(model_path_list)
test_preds = np.argmax(test_preds, axis=-1)
test_names_ds = get_dataset(files_path)
image_names = [img_name.numpy().decode('utf-8') for img, img_name in iter(test_names_ds.unbatch())]
submission = pd.DataFrame({'image_id': image_names, 'label': test_preds})
submission.to_csv('submission.csv', index=False)
display(submission.head())
```
|
github_jupyter
|
!pip install --quiet /kaggle/input/kerasapplications
!pip install --quiet /kaggle/input/efficientnet-git
import warnings, glob
from tensorflow.keras import Sequential, Model
import efficientnet.tfkeras as efn
from cassava_scripts import *
seed = 0
seed_everything(seed)
warnings.filterwarnings('ignore')
# TPU or GPU detection
# Detect hardware, return appropriate distribution strategy
strategy, tpu = set_up_strategy()
AUTO = tf.data.experimental.AUTOTUNE
REPLICAS = strategy.num_replicas_in_sync
print(f'REPLICAS: {REPLICAS}')
BATCH_SIZE = 8 * REPLICAS
HEIGHT = 512
WIDTH = 512
CHANNELS = 3
N_CLASSES = 5
TTA_STEPS = 0 # Do TTA if > 0
def data_augment(image, label):
p_spatial = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_rotate = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_1 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_2 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_3 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_crop = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
# Flips
image = tf.image.random_flip_left_right(image)
image = tf.image.random_flip_up_down(image)
if p_spatial > .75:
image = tf.image.transpose(image)
# Rotates
if p_rotate > .75:
image = tf.image.rot90(image, k=3) # rotate 270º
elif p_rotate > .5:
image = tf.image.rot90(image, k=2) # rotate 180º
elif p_rotate > .25:
image = tf.image.rot90(image, k=1) # rotate 90º
# Pixel-level transforms
if p_pixel_1 >= .4:
image = tf.image.random_saturation(image, lower=.7, upper=1.3)
if p_pixel_2 >= .4:
image = tf.image.random_contrast(image, lower=.8, upper=1.2)
if p_pixel_3 >= .4:
image = tf.image.random_brightness(image, max_delta=.1)
# Crops
if p_crop > .7:
if p_crop > .9:
image = tf.image.central_crop(image, central_fraction=.7)
elif p_crop > .8:
image = tf.image.central_crop(image, central_fraction=.8)
else:
image = tf.image.central_crop(image, central_fraction=.9)
elif p_crop > .4:
crop_size = tf.random.uniform([], int(HEIGHT*.8), HEIGHT, dtype=tf.int32)
image = tf.image.random_crop(image, size=[crop_size, crop_size, CHANNELS])
# # Crops
# if p_crop > .6:
# if p_crop > .9:
# image = tf.image.central_crop(image, central_fraction=.5)
# elif p_crop > .8:
# image = tf.image.central_crop(image, central_fraction=.6)
# elif p_crop > .7:
# image = tf.image.central_crop(image, central_fraction=.7)
# else:
# image = tf.image.central_crop(image, central_fraction=.8)
# elif p_crop > .3:
# crop_size = tf.random.uniform([], int(HEIGHT*.6), HEIGHT, dtype=tf.int32)
# image = tf.image.random_crop(image, size=[crop_size, crop_size, CHANNELS])
return image, label
# Datasets utility functions
def resize_image(image, label):
image = tf.image.resize(image, [HEIGHT, WIDTH])
image = tf.reshape(image, [HEIGHT, WIDTH, CHANNELS])
return image, label
def process_path(file_path):
name = get_name(file_path)
img = tf.io.read_file(file_path)
img = decode_image(img)
img, _ = scale_image(img, None)
# img = center_crop(img, HEIGHT, WIDTH)
return img, name
def get_dataset(files_path, shuffled=False, tta=False, extension='jpg'):
dataset = tf.data.Dataset.list_files(f'{files_path}*{extension}', shuffle=shuffled)
dataset = dataset.map(process_path, num_parallel_calls=AUTO)
if tta:
dataset = dataset.map(data_augment, num_parallel_calls=AUTO)
dataset = dataset.map(resize_image, num_parallel_calls=AUTO)
dataset = dataset.batch(BATCH_SIZE)
dataset = dataset.prefetch(AUTO)
return dataset
database_base_path = '/kaggle/input/cassava-leaf-disease-classification/'
submission = pd.read_csv(f'{database_base_path}sample_submission.csv')
display(submission.head())
TEST_FILENAMES = tf.io.gfile.glob(f'{database_base_path}test_tfrecords/ld_test*.tfrec')
NUM_TEST_IMAGES = count_data_items(TEST_FILENAMES)
print(f'GCS: test: {NUM_TEST_IMAGES}')
model_path_list = glob.glob('/kaggle/input/44-cassava-leaf-effnetb5-2020-sing-cut-out-512x512/*.h5')
model_path_list.sort()
print('Models to predict:')
print(*model_path_list, sep='\n')
def model_fn(input_shape, N_CLASSES):
inputs = L.Input(shape=input_shape, name='input_image')
base_model = efn.EfficientNetB5(input_tensor=inputs,
include_top=False,
weights=None,
pooling='avg')
x = L.Dropout(.25)(base_model.output)
output = L.Dense(N_CLASSES, activation='softmax', name='output')(x)
model = Model(inputs=inputs, outputs=output)
return model
with strategy.scope():
model = model_fn((None, None, CHANNELS), N_CLASSES)
model.summary()
files_path = f'{database_base_path}test_images/'
test_size = len(os.listdir(files_path))
test_preds = np.zeros((test_size, N_CLASSES))
for model_path in model_path_list:
print(model_path)
K.clear_session()
model.load_weights(model_path)
if TTA_STEPS > 0:
test_ds = get_dataset(files_path, tta=True).repeat()
ct_steps = TTA_STEPS * ((test_size/BATCH_SIZE) + 1)
preds = model.predict(test_ds, steps=ct_steps, verbose=1)[:(test_size * TTA_STEPS)]
preds = np.mean(preds.reshape(test_size, TTA_STEPS, N_CLASSES, order='F'), axis=1)
test_preds += preds / len(model_path_list)
else:
test_ds = get_dataset(files_path, tta=False)
x_test = test_ds.map(lambda image, image_name: image)
test_preds += model.predict(x_test) / len(model_path_list)
test_preds = np.argmax(test_preds, axis=-1)
test_names_ds = get_dataset(files_path)
image_names = [img_name.numpy().decode('utf-8') for img, img_name in iter(test_names_ds.unbatch())]
submission = pd.DataFrame({'image_id': image_names, 'label': test_preds})
submission.to_csv('submission.csv', index=False)
display(submission.head())
| 0.5564 | 0.547525 |
<center> <font size='6' font-weight='bold'> Death Certificates (INSEE) - Analysis </font> </center>
<center> <i> Tony WU </i> </center>
The data at hand is given in several .csv files that are named with the following pattern:
`DC_{year}_det.csv` where {Year} is a 4-digit value.
<img src=../ressources/Death_Certificates_INSEE_screen_files.png>
```
import os
import numpy as np
import pandas as pd
import datetime
import seaborn as sns
import matplotlib.pyplot as plt
import geopandas as gpd
import geoplot as gplt
from pprint import pprint
```
# Importing the dataset
```
dir_data = '../data/Death_Certificates_INSEE'
list_csv = os.listdir(dir_data)
list_csv
df_19 = pd.read_csv(os.path.join(dir_data, 'DC_2019_det.csv'), sep=';')
df_20 = pd.read_csv(os.path.join(dir_data, 'DC_2020_det.csv'), sep=';')
df_19.shape, df_20.shape
df_19.head()
df_20.head()
df_19.dtypes
```
# Data cleaning
## Concatenating the two DataFrames
First of all, we'll concatenate the two DataFrames.
```
df = pd.concat([df_19, df_20])
df.head()
len(df_19), len(df_20), len(df)
```
## Cleaning
We notice that `MNAIS`and `JNAIS` are float64 while they are supposed to be int64.
Before converting those columns's types, let's check for potential NANs.
```
df.isna().sum()
```
As we thought, there are quite a lot of NANs.
What's interesting is that even though we sometimes don't have access to the month and day of birth, we always have the year. Thus, instead of getting rid of them, we'll set their birth day to the middle of the year ie the `2nd of July`.
```
# Getting indexes where row contains at least one NAN
df.isna().any(axis=1)
df[df.isna().any(axis=1)]
l_idx_to_change = df.isna().any(axis=1)
df.loc[df.isna().any(axis=1), 'MNAIS'] = 6
df.loc[df.isna().any(axis=1), 'JNAIS'] = 2
df.loc[l_idx_to_change, :].head()
df.isna().sum()
# Changing dtypes to int:
df['MNAIS'] = df['MNAIS'].astype(int)
df['JNAIS'] = df['JNAIS'].astype(int)
df.dtypes
```
We can see that some rows have a wrong birth day. We'll just drop those since there are almost no such rows.
```
df[df['MNAIS']==0].head()
df[df['JNAIS']==0].head()
df = df.drop(df[(df['JNAIS']==0) | (df['MNAIS']==0)].index)
df = df.drop(df[(df['JDEC']==0) | (df['MDEC']==0)].index)
df[df['MNAIS']==0].head()
```
## Converting to datetime64
To have a better workflow, we'll use 1 feature as `datetime64` for both birthday and death day.
In other terms, we'll add two new features `NAIS` (birthday) and `DEC` (death day) and drop all previous features used to give the date (ie year, month and day).
```
df['NAIS'] = pd.to_datetime(dict(year=df['ANAIS'], month=df['MNAIS'], day=df['JNAIS']))
df['DEC'] = pd.to_datetime(dict(year=df['ADEC'], month=df['MDEC'], day=df['JDEC']))
df = df.drop(columns=['ANAIS', 'MNAIS', 'JNAIS', 'ADEC', 'MDEC', 'JDEC'])
df.head()
```
## Feature engineering
It seems interesting to know the age of the person when he/she died.
Thus we'll add such a feature in the DataFrame. We'll call it `AGE_MORT`.
```
test = df['DEC'] - df['NAIS']
test
df['AGE_MORT'] = ((df['DEC'] - df['NAIS']).apply(lambda x: x.days) / 365.25)
df.head(10)
```
# Data analysis
## Feature description
Below is the description of all the given features.
```
features_doc = pd.read_csv(os.path.join(dir_data, 'metadonnees_deces_ficdet.csv'), sep=';')
features_doc
```
**NB: There is a mistake in the given documentation since `NAISS` should be written `NAIS`!**
## Unique values
List of columns:
```
df.columns
df.head()
```
Let's take a look at all the different values for some columns:
```
df['DEPDEC'].unique()
df['COMDEC'].unique()
df['COMDOM'].unique()
len(df['COMDEC'].unique()), len(df['COMDOM'].unique())
df['SEXE'].unique()
df['LIEUDEC2'].unique()
```
## Counting bins with respect to death age
```
bins = [i for i in range(0, 101, 10)]
bins
groups = df['AGE_MORT'].groupby(pd.cut(df['AGE_MORT'], bins))
groups
groups.count()
groups.count().plot.bar(figsize=(10,6))
plt.title("Nombre de décès en fonction des tranches d'âge entre début 2019 et fin 2020")
```
Without the shadow of a doubt, we can see that the older, the more chance to die.
However, we have to make sure that those deaths are COVID-related...
## Searching corrolation between features
```
df.groupby('LIEUDEC2').mean()
```
There is for sure corrolation between the place of death and the death age. However, we still have to make sure that this is a relation of causality...
```
df.groupby('DEPDEC').mean()
```
# GeoPandas
`GeoPandas` comes with three pre-made maps that are already on your computer if you installed the package. Once loaded as `GeoDataFrame`, these maps contain the Points for several major cities or low-resolution Polygons for the borders of all countries.
## First steps
```
# Returns names of available maps
gpd.datasets.available
# Returns path of a particular map
gpd.datasets.get_path('naturalearth_lowres')
# Opens the map as a GeoDataFrame
world = gpd.read_file(gpd.datasets.get_path('naturalearth_lowres'))
world
gplt.polyplot(world, figsize=(10, 8))
```
## Getting France geojson
```
france = gpd.read_file('../data/departements-avec-outre-mer.geojson')
france.head()
france.dtypes
france.code.unique()
gplt.polyplot(france, figsize=(10, 8))
```
Let's try to create a GeoDataFrame containing only the French metropole.
```
france['code'].isin([str(i) for i in range(0, 96)])
metropole = france[france['code'].isin([f'{i:02d}' for i in range(0, 96)])]
metropole
gplt.polyplot(metropole.geometry)
gplt.polyplot(metropole[metropole.nom=='Ain'].geometry)
```
## Joining the two DataFrames
Renaming the department code in both DataFrames to prepare for join:
```
france = france.rename(columns={'code': 'DEPDEC'})
france.head()
```
Let's check that the `inner join` operation won't encounter any error by looking at all the unique values of departments in both DataFrames.
```
np.sort(df.DEPDEC.unique())
np.sort(france['DEPDEC'].unique())
np.all(np.sort(df.DEPDEC.unique()) == np.sort(france['DEPDEC'].unique()))
```
It all seems ok, so let's merge now.
```
df = pd.merge(df, france, on='DEPDEC', how='inner')
df.head()
```
Now let's recompute the death age mean grouped by department.
```
df.groupby('DEPDEC').mean()
df[['DEPDEC', 'geometry']].drop_duplicates(subset=['DEPDEC'])
deaths_by_department = pd.merge(
df.groupby('DEPDEC').agg(['mean', 'count']),
df[['DEPDEC', 'geometry']].drop_duplicates(subset=['DEPDEC']),
on='DEPDEC',
how='inner',
left_index=True
).set_index('DEPDEC')
deaths_by_department = deaths_by_department.rename(
columns={('AGE_MORT', 'mean'): 'mean_death_age', ('AGE_MORT', 'count'): 'death_count'}
)
deaths_by_department
deaths_by_department.geometry
gplt.polyplot(france)
```
We notice that we have to convert the `DataFrame` to `GeoDataFrame` in order to work with it in `GeoPanda`.
```
type(deaths_by_department)
deaths_by_department = gpd.GeoDataFrame(deaths_by_department, geometry=deaths_by_department.geometry)
deaths_by_department
gplt.choropleth(deaths_by_department, hue='mean_death_age')
deaths_by_department_metropole = deaths_by_department[deaths_by_department.index.isin([f'{i:02d}' for i in range(0, 96)])]
deaths_by_department_metropole
gplt.choropleth(deaths_by_department_metropole, hue='mean_death_age', cmap='RdYlGn',legend=True, figsize=(8, 5))
plt.title('Mean death age between of deaths by department in France,\nfrom early 2019 to late 2020');
gplt.choropleth(deaths_by_department_metropole, hue='death_count', cmap='hot_r', legend=True, figsize=(8, 5))
plt.title('Number of deaths between of deaths by department in France,\nfrom early 2019 to late 2020');
```
# Age-Adjusted Rates
According to https://health.mo.gov/data/mica/CDP_MICA/AARate.html, it seems reasonable to consider the use of `Age-Adjusted Rates` instead of death ages to take into account the variability in terms of age distribution across the country.
Here is a definiton given on the website:
> **Age-Adjusted Rates**
Age adjusting rates is a way to make fairer comparisons between groups with different age distributions. For example, a county having a higher percentage of elderly people may have a higher rate of death or hospitalization than a county with a younger population, merely because the elderly are more likely to die or be hospitalized. (The same distortion can happen when comparing races, genders, or time periods.) Age adjustment can make the different groups more comparable.
A "standard" population distribution is used to adjust death and hospitalization rates. The age-adjusted rates are rates that would have existed if the population under study had the same age distribution as the "standard" population. Therefore, they are summary measures adjusted for differences in age distributions.
The National Center for Health Statistics recommends that the U.S. 2000 standard population be used when calculating age-adjusted rates. Users of Missouri Information for Community Assessment (MICA) have the option of selecting age-adjusted rates based on the U.S. 1940, 1970 or 2000 standard populations when generating tables where age-adjustment is utilized. The National Center for Health Statistics recommends that the U.S. 2000 standard population be used when calculating age-adjusted rates. However, if you compare rates from different sources, it is very important that you use the same standard population on both sides of your comparison. It is not legitimate to compare adjusted rates which use different standard populations.
Age-adjusted rates in the Community Data Profiles use the U.S. 2000 standard population.
|
github_jupyter
|
import os
import numpy as np
import pandas as pd
import datetime
import seaborn as sns
import matplotlib.pyplot as plt
import geopandas as gpd
import geoplot as gplt
from pprint import pprint
dir_data = '../data/Death_Certificates_INSEE'
list_csv = os.listdir(dir_data)
list_csv
df_19 = pd.read_csv(os.path.join(dir_data, 'DC_2019_det.csv'), sep=';')
df_20 = pd.read_csv(os.path.join(dir_data, 'DC_2020_det.csv'), sep=';')
df_19.shape, df_20.shape
df_19.head()
df_20.head()
df_19.dtypes
df = pd.concat([df_19, df_20])
df.head()
len(df_19), len(df_20), len(df)
df.isna().sum()
# Getting indexes where row contains at least one NAN
df.isna().any(axis=1)
df[df.isna().any(axis=1)]
l_idx_to_change = df.isna().any(axis=1)
df.loc[df.isna().any(axis=1), 'MNAIS'] = 6
df.loc[df.isna().any(axis=1), 'JNAIS'] = 2
df.loc[l_idx_to_change, :].head()
df.isna().sum()
# Changing dtypes to int:
df['MNAIS'] = df['MNAIS'].astype(int)
df['JNAIS'] = df['JNAIS'].astype(int)
df.dtypes
df[df['MNAIS']==0].head()
df[df['JNAIS']==0].head()
df = df.drop(df[(df['JNAIS']==0) | (df['MNAIS']==0)].index)
df = df.drop(df[(df['JDEC']==0) | (df['MDEC']==0)].index)
df[df['MNAIS']==0].head()
df['NAIS'] = pd.to_datetime(dict(year=df['ANAIS'], month=df['MNAIS'], day=df['JNAIS']))
df['DEC'] = pd.to_datetime(dict(year=df['ADEC'], month=df['MDEC'], day=df['JDEC']))
df = df.drop(columns=['ANAIS', 'MNAIS', 'JNAIS', 'ADEC', 'MDEC', 'JDEC'])
df.head()
test = df['DEC'] - df['NAIS']
test
df['AGE_MORT'] = ((df['DEC'] - df['NAIS']).apply(lambda x: x.days) / 365.25)
df.head(10)
features_doc = pd.read_csv(os.path.join(dir_data, 'metadonnees_deces_ficdet.csv'), sep=';')
features_doc
df.columns
df.head()
df['DEPDEC'].unique()
df['COMDEC'].unique()
df['COMDOM'].unique()
len(df['COMDEC'].unique()), len(df['COMDOM'].unique())
df['SEXE'].unique()
df['LIEUDEC2'].unique()
bins = [i for i in range(0, 101, 10)]
bins
groups = df['AGE_MORT'].groupby(pd.cut(df['AGE_MORT'], bins))
groups
groups.count()
groups.count().plot.bar(figsize=(10,6))
plt.title("Nombre de décès en fonction des tranches d'âge entre début 2019 et fin 2020")
df.groupby('LIEUDEC2').mean()
df.groupby('DEPDEC').mean()
# Returns names of available maps
gpd.datasets.available
# Returns path of a particular map
gpd.datasets.get_path('naturalearth_lowres')
# Opens the map as a GeoDataFrame
world = gpd.read_file(gpd.datasets.get_path('naturalearth_lowres'))
world
gplt.polyplot(world, figsize=(10, 8))
france = gpd.read_file('../data/departements-avec-outre-mer.geojson')
france.head()
france.dtypes
france.code.unique()
gplt.polyplot(france, figsize=(10, 8))
france['code'].isin([str(i) for i in range(0, 96)])
metropole = france[france['code'].isin([f'{i:02d}' for i in range(0, 96)])]
metropole
gplt.polyplot(metropole.geometry)
gplt.polyplot(metropole[metropole.nom=='Ain'].geometry)
france = france.rename(columns={'code': 'DEPDEC'})
france.head()
np.sort(df.DEPDEC.unique())
np.sort(france['DEPDEC'].unique())
np.all(np.sort(df.DEPDEC.unique()) == np.sort(france['DEPDEC'].unique()))
df = pd.merge(df, france, on='DEPDEC', how='inner')
df.head()
df.groupby('DEPDEC').mean()
df[['DEPDEC', 'geometry']].drop_duplicates(subset=['DEPDEC'])
deaths_by_department = pd.merge(
df.groupby('DEPDEC').agg(['mean', 'count']),
df[['DEPDEC', 'geometry']].drop_duplicates(subset=['DEPDEC']),
on='DEPDEC',
how='inner',
left_index=True
).set_index('DEPDEC')
deaths_by_department = deaths_by_department.rename(
columns={('AGE_MORT', 'mean'): 'mean_death_age', ('AGE_MORT', 'count'): 'death_count'}
)
deaths_by_department
deaths_by_department.geometry
gplt.polyplot(france)
type(deaths_by_department)
deaths_by_department = gpd.GeoDataFrame(deaths_by_department, geometry=deaths_by_department.geometry)
deaths_by_department
gplt.choropleth(deaths_by_department, hue='mean_death_age')
deaths_by_department_metropole = deaths_by_department[deaths_by_department.index.isin([f'{i:02d}' for i in range(0, 96)])]
deaths_by_department_metropole
gplt.choropleth(deaths_by_department_metropole, hue='mean_death_age', cmap='RdYlGn',legend=True, figsize=(8, 5))
plt.title('Mean death age between of deaths by department in France,\nfrom early 2019 to late 2020');
gplt.choropleth(deaths_by_department_metropole, hue='death_count', cmap='hot_r', legend=True, figsize=(8, 5))
plt.title('Number of deaths between of deaths by department in France,\nfrom early 2019 to late 2020');
| 0.385375 | 0.957278 |
# Discretization
---
In this notebook, you will deal with continuous state and action spaces by discretizing them. This will enable you to apply reinforcement learning algorithms that are only designed to work with discrete spaces.
### 1. Import the Necessary Packages
```
import sys
import gym
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# Set plotting options
%matplotlib inline
plt.style.use('ggplot')
np.set_printoptions(precision=3, linewidth=120)
```
### 2. Specify the Environment, and Explore the State and Action Spaces
We'll use [OpenAI Gym](https://gym.openai.com/) environments to test and develop our algorithms. These simulate a variety of classic as well as contemporary reinforcement learning tasks. Let's use an environment that has a continuous state space, but a discrete action space.
```
# Create an environment and set random seed
env = gym.make('MountainCar-v0')
env.seed(505);
```
Run the next code cell to watch a random agent.
```
state = env.reset()
score = 0
for t in range(200):
action = env.action_space.sample()
env.render()
state, reward, done, _ = env.step(action)
score += reward
if done:
break
print('Final score:', score)
env.close()
```
In this notebook, you will train an agent to perform much better! For now, we can explore the state and action spaces, as well as sample them.
```
# Explore state (observation) space
print("State space:", env.observation_space)
print("- low:", env.observation_space.low)
print("- high:", env.observation_space.high)
# Generate some samples from the state space
print("State space samples:")
print(np.array([env.observation_space.sample() for i in range(10)]))
# Explore the action space
print("Action space:", env.action_space)
# Generate some samples from the action space
print("Action space samples:")
print(np.array([env.action_space.sample() for i in range(10)]))
```
### 3. Discretize the State Space with a Uniform Grid
We will discretize the space using a uniformly-spaced grid. Implement the following function to create such a grid, given the lower bounds (`low`), upper bounds (`high`), and number of desired `bins` along each dimension. It should return the split points for each dimension, which will be 1 less than the number of bins.
For instance, if `low = [-1.0, -5.0]`, `high = [1.0, 5.0]`, and `bins = (10, 10)`, then your function should return the following list of 2 NumPy arrays:
```
[array([-0.8, -0.6, -0.4, -0.2, 0.0, 0.2, 0.4, 0.6, 0.8]),
array([-4.0, -3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0, 4.0])]
```
Note that the ends of `low` and `high` are **not** included in these split points. It is assumed that any value below the lowest split point maps to index `0` and any value above the highest split point maps to index `n-1`, where `n` is the number of bins along that dimension.
```
def create_uniform_grid(low, high, bins=(10, 10)):
"""Define a uniformly-spaced grid that can be used to discretize a space.
Parameters
----------
low : array_like
Lower bounds for each dimension of the continuous space.
high : array_like
Upper bounds for each dimension of the continuous space.
bins : tuple
Number of bins along each corresponding dimension.
Returns
-------
grid : list of array_like
A list of arrays containing split points for each dimension.
"""
return [np.histogram_bin_edges(np.linspace(low[x], high[x]), bins[x])[1:-1] for x in range(len(bins))]
low = [-1.0, -5.0]
high = [1.0, 5.0]
create_uniform_grid(low, high) # [test]
```
Now write a function that can convert samples from a continuous space into its equivalent discretized representation, given a grid like the one you created above. You can use the [`numpy.digitize()`](https://docs.scipy.org/doc/numpy-1.9.3/reference/generated/numpy.digitize.html) function for this purpose.
Assume the grid is a list of NumPy arrays containing the following split points:
```
[array([-0.8, -0.6, -0.4, -0.2, 0.0, 0.2, 0.4, 0.6, 0.8]),
array([-4.0, -3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0, 4.0])]
```
Here are some potential samples and their corresponding discretized representations:
```
[-1.0 , -5.0] => [0, 0]
[-0.81, -4.1] => [0, 0]
[-0.8 , -4.0] => [1, 1]
[-0.5 , 0.0] => [2, 5]
[ 0.2 , -1.9] => [6, 3]
[ 0.8 , 4.0] => [9, 9]
[ 0.81, 4.1] => [9, 9]
[ 1.0 , 5.0] => [9, 9]
```
**Note**: There may be one-off differences in binning due to floating-point inaccuracies when samples are close to grid boundaries, but that is alright.
```
def discretize(sample, grid):
"""Discretize a sample as per given grid.
Parameters
----------
sample : array_like
A single sample from the (original) continuous space.
grid : list of array_like
A list of arrays containing split points for each dimension.
Returns
-------
discretized_sample : array_like
A sequence of integers with the same number of dimensions as sample.
"""
return [np.digitize(data, grid[i])for i, data in enumerate(sample)]
# Test with a simple grid and some samples
grid = create_uniform_grid([-1.0, -5.0], [1.0, 5.0])
samples = np.array(
[[-1.0 , -5.0],
[-0.81, -4.1],
[-0.8 , -4.0],
[-0.5 , 0.0],
[ 0.2 , -1.9],
[ 0.8 , 4.0],
[ 0.81, 4.1],
[ 1.0 , 5.0]])
discretized_samples = np.array([discretize(sample, grid) for sample in samples])
print("\nSamples:", repr(samples), sep="\n")
print("\nDiscretized samples:", repr(discretized_samples), sep="\n")
```
### 4. Visualization
It might be helpful to visualize the original and discretized samples to get a sense of how much error you are introducing.
```
import matplotlib.collections as mc
def visualize_samples(samples, discretized_samples, grid, low=None, high=None):
"""Visualize original and discretized samples on a given 2-dimensional grid."""
fig, ax = plt.subplots(figsize=(10, 10))
# Show grid
ax.xaxis.set_major_locator(plt.FixedLocator(grid[0]))
ax.yaxis.set_major_locator(plt.FixedLocator(grid[1]))
ax.grid(True)
# If bounds (low, high) are specified, use them to set axis limits
if low is not None and high is not None:
ax.set_xlim(low[0], high[0])
ax.set_ylim(low[1], high[1])
else:
# Otherwise use first, last grid locations as low, high (for further mapping discretized samples)
low = [splits[0] for splits in grid]
high = [splits[-1] for splits in grid]
# Map each discretized sample (which is really an index) to the center of corresponding grid cell
grid_extended = np.hstack((np.array([low]).T, grid, np.array([high]).T)) # add low and high ends
grid_centers = (grid_extended[:, 1:] + grid_extended[:, :-1]) / 2 # compute center of each grid cell
locs = np.stack(grid_centers[i, discretized_samples[:, i]] for i in range(len(grid))).T # map discretized samples
ax.plot(samples[:, 0], samples[:, 1], 'o') # plot original samples
ax.plot(locs[:, 0], locs[:, 1], 's') # plot discretized samples in mapped locations
ax.add_collection(mc.LineCollection(list(zip(samples, locs)), colors='orange')) # add a line connecting each original-discretized sample
ax.legend(['original', 'discretized'])
visualize_samples(samples, discretized_samples, grid, low, high)
```
Now that we have a way to discretize a state space, let's apply it to our reinforcement learning environment.
```
# Create a grid to discretize the state space
state_grid = create_uniform_grid(env.observation_space.low, env.observation_space.high, bins=(10, 10))
state_grid
# Obtain some samples from the space, discretize them, and then visualize them
state_samples = np.array([env.observation_space.sample() for i in range(10)])
discretized_state_samples = np.array([discretize(sample, state_grid) for sample in state_samples])
visualize_samples(state_samples, discretized_state_samples, state_grid,
env.observation_space.low, env.observation_space.high)
plt.xlabel('position'); plt.ylabel('velocity'); # axis labels for MountainCar-v0 state space
```
You might notice that if you have enough bins, the discretization doesn't introduce too much error into your representation. So we may be able to now apply a reinforcement learning algorithm (like Q-Learning) that operates on discrete spaces. Give it a shot to see how well it works!
### 5. Q-Learning
Provided below is a simple Q-Learning agent. Implement the `preprocess_state()` method to convert each continuous state sample to its corresponding discretized representation.
```
class QLearningAgent:
"""Q-Learning agent that can act on a continuous state space by discretizing it."""
def __init__(self, env, state_grid, alpha=0.02, gamma=0.99,
epsilon=1.0, epsilon_decay_rate=0.9995, min_epsilon=.01, seed=505):
"""Initialize variables, create grid for discretization."""
# Environment info
self.env = env
self.state_grid = state_grid
self.state_size = tuple(len(splits) + 1 for splits in self.state_grid) # n-dimensional state space
self.action_size = self.env.action_space.n # 1-dimensional discrete action space
self.seed = np.random.seed(seed)
print("Environment:", self.env)
print("State space size:", self.state_size)
print("Action space size:", self.action_size)
# Learning parameters
self.alpha = alpha # learning rate
self.gamma = gamma # discount factor
self.epsilon = self.initial_epsilon = epsilon # initial exploration rate
self.epsilon_decay_rate = epsilon_decay_rate # how quickly should we decrease epsilon
self.min_epsilon = min_epsilon
# Create Q-table
self.q_table = np.zeros(shape=(self.state_size + (self.action_size,)))
print("Q table size:", self.q_table.shape)
def preprocess_state(self, state):
"""Map a continuous state to its discretized representation."""
return tuple(discretize(state, self.state_grid))
def reset_episode(self, state):
"""Reset variables for a new episode."""
# Gradually decrease exploration rate
self.epsilon *= self.epsilon_decay_rate
self.epsilon = max(self.epsilon, self.min_epsilon)
# Decide initial action
self.last_state = self.preprocess_state(state)
self.last_action = np.argmax(self.q_table[self.last_state])
return self.last_action
def reset_exploration(self, epsilon=None):
"""Reset exploration rate used when training."""
self.epsilon = epsilon if epsilon is not None else self.initial_epsilon
def act(self, state, reward=None, done=None, mode='train'):
"""Pick next action and update internal Q table (when mode != 'test')."""
state = self.preprocess_state(state)
if mode == 'test':
# Test mode: Simply produce an action
action = np.argmax(self.q_table[state])
else:
# Train mode (default): Update Q table, pick next action
# Note: We update the Q table entry for the *last* (state, action) pair with current state, reward
self.q_table[self.last_state + (self.last_action,)] += self.alpha * \
(reward + self.gamma * max(self.q_table[state]) - self.q_table[self.last_state + (self.last_action,)])
# Exploration vs. exploitation
do_exploration = np.random.uniform(0, 1) < self.epsilon
if do_exploration:
# Pick a random action
action = np.random.randint(0, self.action_size)
else:
# Pick the best action from Q table
action = np.argmax(self.q_table[state])
# Roll over current state, action for next step
self.last_state = state
self.last_action = action
return action
q_agent = QLearningAgent(env, state_grid)
```
Let's also define a convenience function to run an agent on a given environment. When calling this function, you can pass in `mode='test'` to tell the agent not to learn.
```
def run(agent, env, num_episodes=20000, mode='train'):
"""Run agent in given reinforcement learning environment and return scores."""
scores = []
max_avg_score = -np.inf
for i_episode in range(1, num_episodes+1):
# Initialize episode
state = env.reset()
action = agent.reset_episode(state)
total_reward = 0
done = False
# Roll out steps until done
while not done:
state, reward, done, info = env.step(action)
total_reward += reward
action = agent.act(state, reward, done, mode)
# Save final score
scores.append(total_reward)
# Print episode stats
if mode == 'train':
if len(scores) > 100:
avg_score = np.mean(scores[-100:])
if avg_score > max_avg_score:
max_avg_score = avg_score
if i_episode % 100 == 0:
print("\rEpisode {}/{} | Max Average Score: {}".format(i_episode, num_episodes, max_avg_score), end="")
sys.stdout.flush()
return scores
scores = run(q_agent, env)
```
The best way to analyze if your agent was learning the task is to plot the scores. It should generally increase as the agent goes through more episodes.
```
# Plot scores obtained per episode
plt.plot(scores); plt.title("Scores");
```
If the scores are noisy, it might be difficult to tell whether your agent is actually learning. To find the underlying trend, you may want to plot a rolling mean of the scores. Let's write a convenience function to plot both raw scores as well as a rolling mean.
```
def plot_scores(scores, rolling_window=100):
"""Plot scores and optional rolling mean using specified window."""
plt.plot(scores); plt.title("Scores");
rolling_mean = pd.Series(scores).rolling(rolling_window).mean()
plt.plot(rolling_mean);
return rolling_mean
rolling_mean = plot_scores(scores)
```
You should observe the mean episode scores go up over time. Next, you can freeze learning and run the agent in test mode to see how well it performs.
```
# Run in test mode and analyze scores obtained
test_scores = run(q_agent, env, num_episodes=100, mode='test')
print("[TEST] Completed {} episodes with avg. score = {}".format(len(test_scores), np.mean(test_scores)))
_ = plot_scores(test_scores, rolling_window=10)
```
It's also interesting to look at the final Q-table that is learned by the agent. Note that the Q-table is of size MxNxA, where (M, N) is the size of the state space, and A is the size of the action space. We are interested in the maximum Q-value for each state, and the corresponding (best) action associated with that value.
```
def plot_q_table(q_table):
"""Visualize max Q-value for each state and corresponding action."""
q_image = np.max(q_table, axis=2) # max Q-value for each state
q_actions = np.argmax(q_table, axis=2) # best action for each state
fig, ax = plt.subplots(figsize=(10, 10))
cax = ax.imshow(q_image, cmap='jet');
cbar = fig.colorbar(cax)
for x in range(q_image.shape[0]):
for y in range(q_image.shape[1]):
ax.text(x, y, q_actions[x, y], color='white',
horizontalalignment='center', verticalalignment='center')
ax.grid(False)
ax.set_title("Q-table, size: {}".format(q_table.shape))
ax.set_xlabel('position')
ax.set_ylabel('velocity')
plot_q_table(q_agent.q_table)
```
### 6. Modify the Grid
Now it's your turn to play with the grid definition and see what gives you optimal results. Your agent's final performance is likely to get better if you use a finer grid, with more bins per dimension, at the cost of higher model complexity (more parameters to learn).
```
# TODO: Create a new agent with a different state space grid
state_grid_new = create_uniform_grid(env.observation_space.low, env.observation_space.high, bins=(20, 20))
q_agent_new = QLearningAgent(env, state_grid_new)
q_agent_new.scores = [] # initialize a list to store scores for this agent
# Train it over a desired number of episodes and analyze scores
# Note: This cell can be run multiple times, and scores will get accumulated
q_agent_new.scores += run(q_agent_new, env, num_episodes=20000) # accumulate scores
rolling_mean_new = plot_scores(q_agent_new.scores)
# Run in test mode and analyze scores obtained
test_scores = run(q_agent_new, env, num_episodes=100, mode='test')
print("[TEST] Completed {} episodes with avg. score = {}".format(len(test_scores), np.mean(test_scores)))
_ = plot_scores(test_scores)
# Visualize the learned Q-table
plot_q_table(q_agent_new.q_table)
```
### 7. Watch a Smart Agent
```
state = env.reset()
score = 0
for t in range(200):
action = q_agent_new.act(state, mode='test')
env.render()
state, reward, done, _ = env.step(action)
score += reward
if done:
break
print('Final score:', score)
env.close()
```
|
github_jupyter
|
import sys
import gym
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# Set plotting options
%matplotlib inline
plt.style.use('ggplot')
np.set_printoptions(precision=3, linewidth=120)
# Create an environment and set random seed
env = gym.make('MountainCar-v0')
env.seed(505);
state = env.reset()
score = 0
for t in range(200):
action = env.action_space.sample()
env.render()
state, reward, done, _ = env.step(action)
score += reward
if done:
break
print('Final score:', score)
env.close()
# Explore state (observation) space
print("State space:", env.observation_space)
print("- low:", env.observation_space.low)
print("- high:", env.observation_space.high)
# Generate some samples from the state space
print("State space samples:")
print(np.array([env.observation_space.sample() for i in range(10)]))
# Explore the action space
print("Action space:", env.action_space)
# Generate some samples from the action space
print("Action space samples:")
print(np.array([env.action_space.sample() for i in range(10)]))
[array([-0.8, -0.6, -0.4, -0.2, 0.0, 0.2, 0.4, 0.6, 0.8]),
array([-4.0, -3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0, 4.0])]
def create_uniform_grid(low, high, bins=(10, 10)):
"""Define a uniformly-spaced grid that can be used to discretize a space.
Parameters
----------
low : array_like
Lower bounds for each dimension of the continuous space.
high : array_like
Upper bounds for each dimension of the continuous space.
bins : tuple
Number of bins along each corresponding dimension.
Returns
-------
grid : list of array_like
A list of arrays containing split points for each dimension.
"""
return [np.histogram_bin_edges(np.linspace(low[x], high[x]), bins[x])[1:-1] for x in range(len(bins))]
low = [-1.0, -5.0]
high = [1.0, 5.0]
create_uniform_grid(low, high) # [test]
[array([-0.8, -0.6, -0.4, -0.2, 0.0, 0.2, 0.4, 0.6, 0.8]),
array([-4.0, -3.0, -2.0, -1.0, 0.0, 1.0, 2.0, 3.0, 4.0])]
[-1.0 , -5.0] => [0, 0]
[-0.81, -4.1] => [0, 0]
[-0.8 , -4.0] => [1, 1]
[-0.5 , 0.0] => [2, 5]
[ 0.2 , -1.9] => [6, 3]
[ 0.8 , 4.0] => [9, 9]
[ 0.81, 4.1] => [9, 9]
[ 1.0 , 5.0] => [9, 9]
def discretize(sample, grid):
"""Discretize a sample as per given grid.
Parameters
----------
sample : array_like
A single sample from the (original) continuous space.
grid : list of array_like
A list of arrays containing split points for each dimension.
Returns
-------
discretized_sample : array_like
A sequence of integers with the same number of dimensions as sample.
"""
return [np.digitize(data, grid[i])for i, data in enumerate(sample)]
# Test with a simple grid and some samples
grid = create_uniform_grid([-1.0, -5.0], [1.0, 5.0])
samples = np.array(
[[-1.0 , -5.0],
[-0.81, -4.1],
[-0.8 , -4.0],
[-0.5 , 0.0],
[ 0.2 , -1.9],
[ 0.8 , 4.0],
[ 0.81, 4.1],
[ 1.0 , 5.0]])
discretized_samples = np.array([discretize(sample, grid) for sample in samples])
print("\nSamples:", repr(samples), sep="\n")
print("\nDiscretized samples:", repr(discretized_samples), sep="\n")
import matplotlib.collections as mc
def visualize_samples(samples, discretized_samples, grid, low=None, high=None):
"""Visualize original and discretized samples on a given 2-dimensional grid."""
fig, ax = plt.subplots(figsize=(10, 10))
# Show grid
ax.xaxis.set_major_locator(plt.FixedLocator(grid[0]))
ax.yaxis.set_major_locator(plt.FixedLocator(grid[1]))
ax.grid(True)
# If bounds (low, high) are specified, use them to set axis limits
if low is not None and high is not None:
ax.set_xlim(low[0], high[0])
ax.set_ylim(low[1], high[1])
else:
# Otherwise use first, last grid locations as low, high (for further mapping discretized samples)
low = [splits[0] for splits in grid]
high = [splits[-1] for splits in grid]
# Map each discretized sample (which is really an index) to the center of corresponding grid cell
grid_extended = np.hstack((np.array([low]).T, grid, np.array([high]).T)) # add low and high ends
grid_centers = (grid_extended[:, 1:] + grid_extended[:, :-1]) / 2 # compute center of each grid cell
locs = np.stack(grid_centers[i, discretized_samples[:, i]] for i in range(len(grid))).T # map discretized samples
ax.plot(samples[:, 0], samples[:, 1], 'o') # plot original samples
ax.plot(locs[:, 0], locs[:, 1], 's') # plot discretized samples in mapped locations
ax.add_collection(mc.LineCollection(list(zip(samples, locs)), colors='orange')) # add a line connecting each original-discretized sample
ax.legend(['original', 'discretized'])
visualize_samples(samples, discretized_samples, grid, low, high)
# Create a grid to discretize the state space
state_grid = create_uniform_grid(env.observation_space.low, env.observation_space.high, bins=(10, 10))
state_grid
# Obtain some samples from the space, discretize them, and then visualize them
state_samples = np.array([env.observation_space.sample() for i in range(10)])
discretized_state_samples = np.array([discretize(sample, state_grid) for sample in state_samples])
visualize_samples(state_samples, discretized_state_samples, state_grid,
env.observation_space.low, env.observation_space.high)
plt.xlabel('position'); plt.ylabel('velocity'); # axis labels for MountainCar-v0 state space
class QLearningAgent:
"""Q-Learning agent that can act on a continuous state space by discretizing it."""
def __init__(self, env, state_grid, alpha=0.02, gamma=0.99,
epsilon=1.0, epsilon_decay_rate=0.9995, min_epsilon=.01, seed=505):
"""Initialize variables, create grid for discretization."""
# Environment info
self.env = env
self.state_grid = state_grid
self.state_size = tuple(len(splits) + 1 for splits in self.state_grid) # n-dimensional state space
self.action_size = self.env.action_space.n # 1-dimensional discrete action space
self.seed = np.random.seed(seed)
print("Environment:", self.env)
print("State space size:", self.state_size)
print("Action space size:", self.action_size)
# Learning parameters
self.alpha = alpha # learning rate
self.gamma = gamma # discount factor
self.epsilon = self.initial_epsilon = epsilon # initial exploration rate
self.epsilon_decay_rate = epsilon_decay_rate # how quickly should we decrease epsilon
self.min_epsilon = min_epsilon
# Create Q-table
self.q_table = np.zeros(shape=(self.state_size + (self.action_size,)))
print("Q table size:", self.q_table.shape)
def preprocess_state(self, state):
"""Map a continuous state to its discretized representation."""
return tuple(discretize(state, self.state_grid))
def reset_episode(self, state):
"""Reset variables for a new episode."""
# Gradually decrease exploration rate
self.epsilon *= self.epsilon_decay_rate
self.epsilon = max(self.epsilon, self.min_epsilon)
# Decide initial action
self.last_state = self.preprocess_state(state)
self.last_action = np.argmax(self.q_table[self.last_state])
return self.last_action
def reset_exploration(self, epsilon=None):
"""Reset exploration rate used when training."""
self.epsilon = epsilon if epsilon is not None else self.initial_epsilon
def act(self, state, reward=None, done=None, mode='train'):
"""Pick next action and update internal Q table (when mode != 'test')."""
state = self.preprocess_state(state)
if mode == 'test':
# Test mode: Simply produce an action
action = np.argmax(self.q_table[state])
else:
# Train mode (default): Update Q table, pick next action
# Note: We update the Q table entry for the *last* (state, action) pair with current state, reward
self.q_table[self.last_state + (self.last_action,)] += self.alpha * \
(reward + self.gamma * max(self.q_table[state]) - self.q_table[self.last_state + (self.last_action,)])
# Exploration vs. exploitation
do_exploration = np.random.uniform(0, 1) < self.epsilon
if do_exploration:
# Pick a random action
action = np.random.randint(0, self.action_size)
else:
# Pick the best action from Q table
action = np.argmax(self.q_table[state])
# Roll over current state, action for next step
self.last_state = state
self.last_action = action
return action
q_agent = QLearningAgent(env, state_grid)
def run(agent, env, num_episodes=20000, mode='train'):
"""Run agent in given reinforcement learning environment and return scores."""
scores = []
max_avg_score = -np.inf
for i_episode in range(1, num_episodes+1):
# Initialize episode
state = env.reset()
action = agent.reset_episode(state)
total_reward = 0
done = False
# Roll out steps until done
while not done:
state, reward, done, info = env.step(action)
total_reward += reward
action = agent.act(state, reward, done, mode)
# Save final score
scores.append(total_reward)
# Print episode stats
if mode == 'train':
if len(scores) > 100:
avg_score = np.mean(scores[-100:])
if avg_score > max_avg_score:
max_avg_score = avg_score
if i_episode % 100 == 0:
print("\rEpisode {}/{} | Max Average Score: {}".format(i_episode, num_episodes, max_avg_score), end="")
sys.stdout.flush()
return scores
scores = run(q_agent, env)
# Plot scores obtained per episode
plt.plot(scores); plt.title("Scores");
def plot_scores(scores, rolling_window=100):
"""Plot scores and optional rolling mean using specified window."""
plt.plot(scores); plt.title("Scores");
rolling_mean = pd.Series(scores).rolling(rolling_window).mean()
plt.plot(rolling_mean);
return rolling_mean
rolling_mean = plot_scores(scores)
# Run in test mode and analyze scores obtained
test_scores = run(q_agent, env, num_episodes=100, mode='test')
print("[TEST] Completed {} episodes with avg. score = {}".format(len(test_scores), np.mean(test_scores)))
_ = plot_scores(test_scores, rolling_window=10)
def plot_q_table(q_table):
"""Visualize max Q-value for each state and corresponding action."""
q_image = np.max(q_table, axis=2) # max Q-value for each state
q_actions = np.argmax(q_table, axis=2) # best action for each state
fig, ax = plt.subplots(figsize=(10, 10))
cax = ax.imshow(q_image, cmap='jet');
cbar = fig.colorbar(cax)
for x in range(q_image.shape[0]):
for y in range(q_image.shape[1]):
ax.text(x, y, q_actions[x, y], color='white',
horizontalalignment='center', verticalalignment='center')
ax.grid(False)
ax.set_title("Q-table, size: {}".format(q_table.shape))
ax.set_xlabel('position')
ax.set_ylabel('velocity')
plot_q_table(q_agent.q_table)
# TODO: Create a new agent with a different state space grid
state_grid_new = create_uniform_grid(env.observation_space.low, env.observation_space.high, bins=(20, 20))
q_agent_new = QLearningAgent(env, state_grid_new)
q_agent_new.scores = [] # initialize a list to store scores for this agent
# Train it over a desired number of episodes and analyze scores
# Note: This cell can be run multiple times, and scores will get accumulated
q_agent_new.scores += run(q_agent_new, env, num_episodes=20000) # accumulate scores
rolling_mean_new = plot_scores(q_agent_new.scores)
# Run in test mode and analyze scores obtained
test_scores = run(q_agent_new, env, num_episodes=100, mode='test')
print("[TEST] Completed {} episodes with avg. score = {}".format(len(test_scores), np.mean(test_scores)))
_ = plot_scores(test_scores)
# Visualize the learned Q-table
plot_q_table(q_agent_new.q_table)
state = env.reset()
score = 0
for t in range(200):
action = q_agent_new.act(state, mode='test')
env.render()
state, reward, done, _ = env.step(action)
score += reward
if done:
break
print('Final score:', score)
env.close()
| 0.640861 | 0.986258 |
# Tema 08 - Programacion Orientada a Objetos (Enunciados)
*Nota: Estos ejercicios son optativos para hacer al final de la unidad y están pensados para apoyar tu aprendizaje*.
**En este ejercicio vas a trabajar el concepto de puntos, coordenadas y vectores sobre el plano cartesiano y cómo la programación Orientada a Objetos puede ser una excelente aliada para trabajar con ellos. No está pensado para que hagas ningún tipo de cálculo sino para que practiques la automatización de tareas.**
*Nota: Creo que es un ejemplo muy interesante, punto de partida en la programación de gráficos, pero si consideras que esto no lo tuyo puedes simplemente pasar de largo. Ahora bien, debes ser consciente de que te vas a perder uno de los ejercicios más interesantes del curso.*
**Antes de continuar voy a explicar brevemente los conceptos básicos por si alguien necesita un repaso.**
## El plano cartesiano
Representa un espacio bidimensional (en 2 dimensiones), formado por dos rectas perpendiculares, una horizontal y otra vertical que se cortan en un punto. La recta horizontal se denomina eje de las abscisas o **eje X**, mientras que la vertical recibe el nombre de eje de las ordenadas o simplemente **eje Y**. En cuanto al punto donde se cortan, se conoce como el **punto de origen O**.
<img src="http://www.escueladevideojuegos.net/ejemplos_edv/Cursos/Python/eje.jpg" width="350" />
Es importante remarcar que el plano se divide en 4 cuadrantes:
<img src="http://www.escueladevideojuegos.net/ejemplos_edv/Cursos/Python/cuadrante.jpg" width="350" />
## Puntos y coordenadas
El objetivo de todo esto es describir la posición de **puntos** sobre el plano en forma de **coordenadas**, que se forman asociando el valor del eje de las X (horizontal) con el valor del eje Y (vertical).
La representación de un punto es sencilla: **P(X,Y)** dónde X y la Y son la distancia horizontal (izquierda o derecha) y vertical (arriba o abajo) respectivamente, utilizando como referencia el punto de origen (0,0), justo en el centro del plano.
<img src="http://www.escueladevideojuegos.net/ejemplos_edv/Cursos/Python/Cartesian-coordinate-system.svg.png" width="300" />
## Vectores en el plano
Finalmente, un vector en el plano hace referencia a un segmento orientado, generado a partir de dos puntos distintos.
A efectos prácticos no deja de ser una línea formada desde un punto inicial en dirección a otro punto final, por lo que se entiende que un vector tiene longitud y dirección/sentido.
<img src="http://www.escueladevideojuegos.net/ejemplos_edv/Cursos/Python/vector3.png" width="300" />
En esta figura, podemos observar dos puntos A y B que podríamos definir de la siguiente forma:
* **A(x1, y1)** => **A(2, 3)**
* **B(x2, y2)** => **B(5, 5)**
Y el vector se representaría como la diferencia entre las coordendas del segundo punto respecto al primero (el segundo menos el primero):
* **AB = (x2-x1, y2-y1)** => **(5-2, 5-3)** => **(3,2)**
Lo que en definitiva no deja de ser: 3 a la derecha y 2 arriba.
Y con esto finalizamos este mini repaso.
# El ejercicio
#### Preparación
* Crea una clase llamada **Punto** con sus dos coordenadas X e Y.
* Añade un método **constructor** para crear puntos fácilmente. Si no se reciben una coordenada, su valor será cero.
* Sobreescribe el método **string**, para que al imprimir por pantalla un punto aparezca en formato (X,Y)
* Añade un método llamado **cuadrante** que indique a qué cuadrante pertenece el punto, o si es el origen.
* Añade un método llamado **vector**, que tome otro punto y calcule el vector resultante entre los dos puntos.
* (Optativo) Añade un método llamado **distancia**, que tome otro punto y calcule la distancia entre los dos puntos y la muestre por pantalla. La fórmula es la siguiente:
<img src="http://www.escueladevideojuegos.net/ejemplos_edv/Cursos/Python/distancia.png" width="250" />
*Nota: La función raíz cuadrada en Python sqrt() se debe importar del módulo math y utilizarla de la siguiente forma:*
```python
import math
math.sqrt(9)
> 3.0
```
* Crea una clase llamada **Rectangulo** con dos puntos (inicial y final) que formarán la diagonal del rectángulo.
* Añade un método **constructor** para crear ambos puntos fácilmente, si no se envían se crearán dos puntos en el origen por defecto.
* Añade al rectángulo un método llamado **base** que muestre la base.
* Añade al rectángulo un método llamado **altura** que muestre la altura.
* Añade al rectángulo un método llamado **area** que muestre el area.
*Puedes identificar fácilmente estos valores si intentas dibujar el cuadrado a partir de su diagonal. Si andas perdido, prueba de dibujarlo en un papel, ¡seguro que lo verás mucho más claro! Además recuerda que puedes utilizar la función **abs()** para saber el valor absolute de un número.*
#### Experimentación
* Crea los puntos A(2, 3), B(5,5), C(-3, -1) y D(0,0) e imprimelos por pantalla.
* Consulta a que cuadrante pertenecen el punto A, C y D.
* Consulta los vectores AB y BA.
* (Optativo) Consulta la distancia entre los puntos 'A y B' y 'B y A'.
* (Optativo) Determina cual de los 3 puntos A, B o C, se encuentra más lejos del origen, punto (0,0).
* Crea un rectángulo utilizando los puntos A y B.
* Consulta la base, altura y área del rectángulo.
```
# Completa el ejercicio aquí
```
|
github_jupyter
|
import math
math.sqrt(9)
> 3.0
# Completa el ejercicio aquí
| 0.181191 | 0.953837 |
# 5. Open types
Consider again the (modified) example from Section 2, in which eligibility for acceptance into St. John's secondary school was to specified:
```
Fact applicant
Fact primary-school Identified by StMary, StGeorge
Fact gpa Identified by 1..4
Fact diploma Identified by primary-school * applicant * gpa
Fact application Identified by applicant * diploma
Fact accepted Identified by applicant
Act accept-application Related to application
Holds when application
Conditioned by application.diploma && diploma.gpa >= 3
Creates accepted()
.
+applicant(Alice).
+applicant(Bob).
+applicant(Chloe).
+diploma(StMary, Alice, 3).
+diploma(StGeorge, Bob, 3).
+diploma(StGeorge, Chloe, 2).
?Enabled(accept-application(StJohn, application(Alice, diploma(StMary, Alice, 3)))).
?Enabled(accept-application(StJohn, application(Bob, diploma(StGeorge, Bob, 3)))).
?!Enabled(accept-application(StJohn, application(Chloe, diploma(StGeorge, Chloe, 2)))).
```
The previous code fragment checks whether Alice, Bob and/or Chloe can be accepted into the school based on their diplomas and whether they are already considered applicants of the school. These two kinds of facts of the case -- recognised diplomas and being an applicant -- are very different in nature in that the school can be expected to know the latter kind (based on an application letter, for example) but may have to rely on some external authority to determine whether the diplomas are valid and indeed handed out by the corresponding primary-school.
If we imagine an eFLINT service running within the local network of St. John's, then the information about applicants can come from within that network, for example, by triggering the action `send-application-letter`.
```
Act send-application-letter Actor applicant Related to diploma
Holds when !applicant // always enabled for non-applicants
Creates application()
.
send-application-letter(David, diploma(StMary, David, 4))
```
When specifying the policy of St. John's in eFLINT, we can decide we have confidence in the school's application-processing software to reliably send updates about received letters to the eFLINT service (perhaps because both services are under our full control). In these conditions, it is safe to apply the **closed-world assumption** to instances of the type `applicant` -- if the eFLINT services does not know `X` applied, then `X` did indeed not apply.
The same cannot be said for the validity of diplomas and we may wish to verify that the primary-school did indeed issue this exact diploma. To support such situations, types can be declared as being 'open' (with the `Open` modifier, see subsection 1.7).
```
Open Fact diploma Identified by primary-school * applicant * gpa
```
To determine whether a particular instance of an open type holds true, it is checked whether it is stated being as true (or false) within the *input* provided alongside the phrase being executed, then whether it is stated as being true (or false) within the current knowledge base, then whether it can be derived as holding true, and finally, if neither of the aforementioned yielded a truth value, then a "missing input" exception is thrown.
```
?Enabled(accept-application(StJohn, application(Alice, diploma(StMary, Alice, 3)))).
?Enabled(accept-application(StJohn, application(Bob, diploma(StGeorge, Bob, 3)))).
?!Enabled(accept-application(StJohn, application(Chloe, diploma(StGeorge, Chloe, 2)))).
?!Enabled(accept-application(StJohn, application(David, diploma(StMary, David, 4)))).
```
This information feeds back into the execution context (e.g. the eFLINT service) and might trigger the execution of a handler to provide the missing information (e.g. a notification sent to an employee of the school to reach out to primary school for validation).
In the `eflint-repl`, a recursive prompt starts to ask the programmer for any assignments for missing input whenever instances of open types were evaluated during the execution of a phrase. This prompt repeatedly asks the programmer to provide the input for instances of open types that are required to complete the execution of the phrase until all missing input is given.
A future version of the Jupyter Kernel for eFLINT may support one or more approaches to providing input and handling missing input.
Note that the input mechanism can be used to replace the internal knowledge base of eFLINT entirely, effectively creating an external knowledge base (e.g. in the form of a persistent database).
|
github_jupyter
|
Fact applicant
Fact primary-school Identified by StMary, StGeorge
Fact gpa Identified by 1..4
Fact diploma Identified by primary-school * applicant * gpa
Fact application Identified by applicant * diploma
Fact accepted Identified by applicant
Act accept-application Related to application
Holds when application
Conditioned by application.diploma && diploma.gpa >= 3
Creates accepted()
.
+applicant(Alice).
+applicant(Bob).
+applicant(Chloe).
+diploma(StMary, Alice, 3).
+diploma(StGeorge, Bob, 3).
+diploma(StGeorge, Chloe, 2).
?Enabled(accept-application(StJohn, application(Alice, diploma(StMary, Alice, 3)))).
?Enabled(accept-application(StJohn, application(Bob, diploma(StGeorge, Bob, 3)))).
?!Enabled(accept-application(StJohn, application(Chloe, diploma(StGeorge, Chloe, 2)))).
Act send-application-letter Actor applicant Related to diploma
Holds when !applicant // always enabled for non-applicants
Creates application()
.
send-application-letter(David, diploma(StMary, David, 4))
Open Fact diploma Identified by primary-school * applicant * gpa
?Enabled(accept-application(StJohn, application(Alice, diploma(StMary, Alice, 3)))).
?Enabled(accept-application(StJohn, application(Bob, diploma(StGeorge, Bob, 3)))).
?!Enabled(accept-application(StJohn, application(Chloe, diploma(StGeorge, Chloe, 2)))).
?!Enabled(accept-application(StJohn, application(David, diploma(StMary, David, 4)))).
| 0.302494 | 0.957358 |
My data is in a JSON format right now. Let's put it into a form that the PC-GAN notebook/pytorch can read.
# Get raw data
```
import sys
import json
raw_data_folder = 'raw_data/'
with open(raw_data_folder + 'co_data.json', 'r') as file_handle:
co_documents = json.load(file_handle)
with open(raw_data_folder + 'h_data.json', 'r') as file_handle:
h_documents = json.load(file_handle)
documents = {'CO': co_documents, 'H': h_documents}
documents['CO'][0]
```
# Save processed data
```
elements = {'H': 1, 'He': 2, 'Li': 3, 'Be': 4, 'B': 5, 'C': 6, 'N': 7, 'O': 8, 'F': 9, 'Ne': 10,
'Na': 11, 'Mg': 12, 'Al': 13, 'Si': 14, 'P': 15, 'S': 16, 'Cl': 17, 'Ar': 18,
'K': 19, 'Ca': 20, 'Sc': 21, 'Ti': 22, 'V': 23, 'Cr': 24, 'Mn': 25, 'Fe': 26,
'Co': 27, 'Ni': 28, 'Cu': 29, 'Zn': 30, 'Ga': 31, 'Ge': 32, 'As': 33, 'Se': 34,
'Br': 35, 'Kr': 36, 'Rb': 37, 'Sr': 38, 'Y': 39, 'Zr': 40, 'Nb': 41, 'Mo': 42,
'Tc': 43, 'Ru': 44, 'Rh': 45, 'Pd': 46, 'Ag': 47, 'Cd': 48, 'In': 49, 'Sn': 50,
'Sb': 51, 'Te': 52, 'I': 53, 'Xe': 54, 'Cs': 55, 'Ba': 56, 'La': 57, 'Ce': 58,
'Pr': 59, 'Nd': 60, 'Pm': 61, 'Sm': 62, 'Eu': 63, 'Gd': 64, 'Tb': 65, 'Dy': 66,
'Ho': 67, 'Er': 68, 'Tm': 69, 'Yb': 70, 'Lu': 71, 'Hf': 72, 'Ta': 73, 'W': 74,
'Re': 75, 'Os': 76, 'Ir': 77, 'Pt': 78, 'Au': 79, 'Hg': 80, 'Tl': 81, 'Pb': 82,
'Bi': 83, 'Po': 84, 'At': 85, 'Rn': 86, 'Fr': 87, 'Ra': 88, 'Ac': 89, 'Th': 90,
'Pa': 91, 'U': 92, 'Np': 93, 'Pu': 94, 'Am': 95}
data_folder = 'gaspy/data/'
for adsorbate, docs in documents.iteritems():
for doc in docs:
subfolder_prefix = doc['adsorbate']
doc_hash = str(hash(json.dumps(doc, sort_keys=True)) % ((sys.maxsize + 1) * 2))
# Write the relaxed structures
subfolder = subfolder_prefix + '_relaxed/'
pts_fname = data_folder + subfolder + 'points/' + doc_hash + '.pts'
with open(pts_fname, 'w') as file_handle:
for atom in doc['atoms']['atoms']:
file_handle.write(' '.join(map(str, atom['position'])) + '\n')
labels_fname = data_folder + subfolder + 'points_label/' + doc_hash + '.seg'
with open(labels_fname, 'w') as file_handle:
for atom in doc['atoms']['atoms']:
element_num = elements[atom['symbol']]
file_handle.write(str(element_num) + '\n')
# Write the unrelaxed structures
subfolder = subfolder_prefix + '_unrelaxed/'
pts_fname = data_folder + subfolder + 'points/' + doc_hash + '.pts'
with open(pts_fname, 'w') as file_handle:
for atom in doc['initial_configuration']['atoms']['atoms']:
file_handle.write(' '.join(map(str, atom['position'])) + '\n')
labels_fname = data_folder + subfolder + 'points_label/' + doc_hash + '.seg'
with open(labels_fname, 'w') as file_handle:
for atom in doc['initial_configuration']['atoms']['atoms']:
element_num = elements[atom['symbol']]
file_handle.write(str(element_num) + '\n')
```
# View examples
```
import random
from ase import Atoms, Atom
from ase.visualize import view
from ase.constraints import dict2constraint
from ase.calculators.singlepoint import SinglePointCalculator
def make_atoms_from_doc(doc):
'''
This is the inversion function for `make_doc_from_atoms`; it takes
Mongo documents created by that function and turns them back into
an ase.Atoms object.
Args:
doc Dictionary/json/Mongo document created by the
`make_doc_from_atoms` function.
Returns:
atoms ase.Atoms object with an ase.SinglePointCalculator attached
'''
atoms = Atoms([Atom(atom['symbol'],
atom['position'],
tag=atom['tag'],
momentum=atom['momentum'],
magmom=atom['magmom'],
charge=atom['charge'])
for atom in doc['atoms']['atoms']],
cell=doc['atoms']['cell'],
pbc=doc['atoms']['pbc'],
info=doc['atoms']['info'],
constraint=[dict2constraint(constraint_dict)
for constraint_dict in doc['atoms']['constraints']])
results = doc['results']
calc = SinglePointCalculator(energy=results.get('energy', None),
forces=results.get('forces', None),
stress=results.get('stress', None),
atoms=atoms)
atoms.set_calculator(calc)
return atoms
co_doc = random.sample(documents['CO'], 1)[0]
initial_co_atoms = make_atoms_from_doc(co_doc['initial_configuration'])
view(initial_co_atoms, viewer='x3d')
final_co_atoms = make_atoms_from_doc(co_doc)
view(final_co_atoms, viewer='x3d')
h_doc = random.sample(documents['H'], 1)[0]
initial_h_atoms = make_atoms_from_doc(h_doc['initial_configuration'])
view(initial_h_atoms, viewer='x3d')
final_h_atoms = make_atoms_from_doc(h_doc['initial_configuration'])
view(final_h_atoms, viewer='x3d')
```
# View color map
```
atoms = Atoms()
for i, (symbol, _) in enumerate(sorted(elements.iteritems(), key=lambda (k, v): (v, k))):
atom = Atom(symbol, position=[i*5, 0, 0])
atoms.append(atom)
view(atoms, viewer='x3d')
```
|
github_jupyter
|
import sys
import json
raw_data_folder = 'raw_data/'
with open(raw_data_folder + 'co_data.json', 'r') as file_handle:
co_documents = json.load(file_handle)
with open(raw_data_folder + 'h_data.json', 'r') as file_handle:
h_documents = json.load(file_handle)
documents = {'CO': co_documents, 'H': h_documents}
documents['CO'][0]
elements = {'H': 1, 'He': 2, 'Li': 3, 'Be': 4, 'B': 5, 'C': 6, 'N': 7, 'O': 8, 'F': 9, 'Ne': 10,
'Na': 11, 'Mg': 12, 'Al': 13, 'Si': 14, 'P': 15, 'S': 16, 'Cl': 17, 'Ar': 18,
'K': 19, 'Ca': 20, 'Sc': 21, 'Ti': 22, 'V': 23, 'Cr': 24, 'Mn': 25, 'Fe': 26,
'Co': 27, 'Ni': 28, 'Cu': 29, 'Zn': 30, 'Ga': 31, 'Ge': 32, 'As': 33, 'Se': 34,
'Br': 35, 'Kr': 36, 'Rb': 37, 'Sr': 38, 'Y': 39, 'Zr': 40, 'Nb': 41, 'Mo': 42,
'Tc': 43, 'Ru': 44, 'Rh': 45, 'Pd': 46, 'Ag': 47, 'Cd': 48, 'In': 49, 'Sn': 50,
'Sb': 51, 'Te': 52, 'I': 53, 'Xe': 54, 'Cs': 55, 'Ba': 56, 'La': 57, 'Ce': 58,
'Pr': 59, 'Nd': 60, 'Pm': 61, 'Sm': 62, 'Eu': 63, 'Gd': 64, 'Tb': 65, 'Dy': 66,
'Ho': 67, 'Er': 68, 'Tm': 69, 'Yb': 70, 'Lu': 71, 'Hf': 72, 'Ta': 73, 'W': 74,
'Re': 75, 'Os': 76, 'Ir': 77, 'Pt': 78, 'Au': 79, 'Hg': 80, 'Tl': 81, 'Pb': 82,
'Bi': 83, 'Po': 84, 'At': 85, 'Rn': 86, 'Fr': 87, 'Ra': 88, 'Ac': 89, 'Th': 90,
'Pa': 91, 'U': 92, 'Np': 93, 'Pu': 94, 'Am': 95}
data_folder = 'gaspy/data/'
for adsorbate, docs in documents.iteritems():
for doc in docs:
subfolder_prefix = doc['adsorbate']
doc_hash = str(hash(json.dumps(doc, sort_keys=True)) % ((sys.maxsize + 1) * 2))
# Write the relaxed structures
subfolder = subfolder_prefix + '_relaxed/'
pts_fname = data_folder + subfolder + 'points/' + doc_hash + '.pts'
with open(pts_fname, 'w') as file_handle:
for atom in doc['atoms']['atoms']:
file_handle.write(' '.join(map(str, atom['position'])) + '\n')
labels_fname = data_folder + subfolder + 'points_label/' + doc_hash + '.seg'
with open(labels_fname, 'w') as file_handle:
for atom in doc['atoms']['atoms']:
element_num = elements[atom['symbol']]
file_handle.write(str(element_num) + '\n')
# Write the unrelaxed structures
subfolder = subfolder_prefix + '_unrelaxed/'
pts_fname = data_folder + subfolder + 'points/' + doc_hash + '.pts'
with open(pts_fname, 'w') as file_handle:
for atom in doc['initial_configuration']['atoms']['atoms']:
file_handle.write(' '.join(map(str, atom['position'])) + '\n')
labels_fname = data_folder + subfolder + 'points_label/' + doc_hash + '.seg'
with open(labels_fname, 'w') as file_handle:
for atom in doc['initial_configuration']['atoms']['atoms']:
element_num = elements[atom['symbol']]
file_handle.write(str(element_num) + '\n')
import random
from ase import Atoms, Atom
from ase.visualize import view
from ase.constraints import dict2constraint
from ase.calculators.singlepoint import SinglePointCalculator
def make_atoms_from_doc(doc):
'''
This is the inversion function for `make_doc_from_atoms`; it takes
Mongo documents created by that function and turns them back into
an ase.Atoms object.
Args:
doc Dictionary/json/Mongo document created by the
`make_doc_from_atoms` function.
Returns:
atoms ase.Atoms object with an ase.SinglePointCalculator attached
'''
atoms = Atoms([Atom(atom['symbol'],
atom['position'],
tag=atom['tag'],
momentum=atom['momentum'],
magmom=atom['magmom'],
charge=atom['charge'])
for atom in doc['atoms']['atoms']],
cell=doc['atoms']['cell'],
pbc=doc['atoms']['pbc'],
info=doc['atoms']['info'],
constraint=[dict2constraint(constraint_dict)
for constraint_dict in doc['atoms']['constraints']])
results = doc['results']
calc = SinglePointCalculator(energy=results.get('energy', None),
forces=results.get('forces', None),
stress=results.get('stress', None),
atoms=atoms)
atoms.set_calculator(calc)
return atoms
co_doc = random.sample(documents['CO'], 1)[0]
initial_co_atoms = make_atoms_from_doc(co_doc['initial_configuration'])
view(initial_co_atoms, viewer='x3d')
final_co_atoms = make_atoms_from_doc(co_doc)
view(final_co_atoms, viewer='x3d')
h_doc = random.sample(documents['H'], 1)[0]
initial_h_atoms = make_atoms_from_doc(h_doc['initial_configuration'])
view(initial_h_atoms, viewer='x3d')
final_h_atoms = make_atoms_from_doc(h_doc['initial_configuration'])
view(final_h_atoms, viewer='x3d')
atoms = Atoms()
for i, (symbol, _) in enumerate(sorted(elements.iteritems(), key=lambda (k, v): (v, k))):
atom = Atom(symbol, position=[i*5, 0, 0])
atoms.append(atom)
view(atoms, viewer='x3d')
| 0.44746 | 0.801276 |
```
!pip install fastFM
!pip install surprise
!pip install lightfm
!pip install hyperopt
!pip uninstall tensorflow
!pip install tensorflow==1.15
!pip install annoy
```
## Imports
```
import pandas as pd
import math
import numpy as np
import random
from numpy.linalg import inv
from numpy.linalg import multi_dot
import matplotlib.pyplot as plt
import itertools
import warnings
from math import sqrt
from sklearn.feature_extraction import DictVectorizer
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder
from fastFM import als
from sklearn.metrics import mean_squared_error
from lightfm.datasets import fetch_movielens
from lightfm import LightFM
from pydrive.auth import GoogleAuth
from pydrive.drive import GoogleDrive
from google.colab import auth
from oauth2client.client import GoogleCredentials
warnings.filterwarnings("ignore")
from scipy import sparse
from lightfm import LightFM
from sklearn.metrics.pairwise import cosine_similarity
from hyperopt import tpe, fmin, hp, Trials, STATUS_OK,space_eval
import sys
import seaborn as sns
from timeit import Timer
from datetime import datetime
from hyperopt import tpe, fmin, hp, Trials, STATUS_OK,space_eval
from collections import defaultdict
import random
from sklearn.metrics import mean_squared_error
from numpy.linalg import inv
import pandas as pd
import numpy as np
import math
from numpy.linalg import inv
from numpy.linalg import multi_dot
import matplotlib.pyplot as plt
import itertools
import warnings
warnings.filterwarnings("ignore")
from annoy import AnnoyIndex
from ncf_singlenode import *
from dataset import Dataset as NCFDataset
from constants import SEED as DEFAULT_SEED
SEED = DEFAULT_SEED
```
## Get data
```
def get_data():
!pip install -U -q PyDrive
from pydrive.auth import GoogleAuth
from pydrive.drive import GoogleDrive
from google.colab import auth
from oauth2client.client import GoogleCredentials
# Authenticate and create the PyDrive client.
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
id = "1FFYnZRIuzQLeBkDUuJpK_oRtUmdxMd9O"
downloaded = drive.CreateFile({'id':id})
downloaded.GetContentFile('final_dataset.csv')
data = pd.read_csv('final_dataset.csv')
return data
data = get_data()
def train_test_split(data):
user_freq=data.groupby(['userId']).size().reset_index(name='counts')
users_lt3=user_freq[user_freq['counts']<3][['userId']]
users_ge3=user_freq[user_freq['counts']>=3][['userId']]
train1=pd.merge(data, users_lt3, on=['userId'],how='inner')
data1=pd.merge(data, users_ge3, on=['userId'],how='inner')
data1.sort_values(['userId', 'timestamp'], ascending=[True, False],inplace=True)
test=data1.groupby('userId').sample(frac=.3, random_state=2)
test_idx=data1.index.isin(test.index.to_list())
train=train1.append(data1[~test_idx])
return train, test, user_freq
train, test, user_freq = train_test_split(data)
```
## ANN-NCF model
```
def run_ann_ncf_pipeline(train, test, user_freq):
train_df = train
test_df = test
def create_SVD_UV(train,test,user_freq):
train=train[['userId', 'movieId', 'rating']]
test=test[['userId', 'movieId', 'rating']]
reader = Reader(rating_scale=(1,5))
train = Dataset.load_from_df(train[['userId', 'movieId', 'rating']], reader=reader)
test = Dataset.load_from_df(test[['userId', 'movieId', 'rating']], reader=reader)
raw_ratings = test.raw_ratings
threshold = int(1 * len(raw_ratings))
A_raw_ratings = raw_ratings[:threshold]
test = test.construct_testset(A_raw_ratings)
raw_ratings1 = train.raw_ratings
threshold = int(1 * len(raw_ratings1))
B_raw_ratings = raw_ratings1[:threshold]
train_test = train.construct_testset(B_raw_ratings)
model = SVD(n_epochs=50,n_factors=15,reg_all=0.1,lr_all=0.02)
trainset = train.build_full_trainset()
model.fit(trainset)
# Retrieiving inner ids, as used by surprise package during model training
user_inner_ids = [x for x in trainset.all_users()]
item_inner_ids = [i for i in trainset.all_items()]
# All ids mapped back to values in the actual train set
user_raw_ids = [trainset.to_raw_uid(x) for x in user_inner_ids]
item_raw_ids = [trainset.to_raw_iid(x) for x in item_inner_ids]
U = model.pu
V = model.qi
return U, V, trainset, user_inner_ids
U,V, trainset, user_inner_ids = create_SVD_UV(train,test,user_freq)
def run_ann(user_inner_ids, V, train_df):
#To be used for serialized process; user item search
f = 15 # n_factors
t = AnnoyIndex(f, 'angular') # Length of item vector that will be indexed
for i,v in zip(user_inner_ids,V):
t.add_item(i, v)
t.build(100)
t.save('test_user_item.ann')
u = AnnoyIndex(f, 'angular')
u.load('test_user_item.ann')
# super fast, will just map the file
user_ids_nn = list(user_freq[user_freq['num_movies']>50]['userId'].values)
def find_nn_greater_than_k(user_id, k=100):
return t.get_nns_by_vector(U[trainset.to_inner_uid(user_id)], k)
user_nn_items = list(map(find_nn_greater_than_k,user_ids_nn))
user_nn_items_dict={}
for userid,items in zip(user_ids_nn,user_nn_items):
user_nn_items_dict[userid] = [trainset.to_raw_iid(i) for i in items]
user_nn_items_df = pd.concat({k: pd.Series(v) for k, v in user_nn_items_dict.items()})
user_nn_items_df = user_nn_items_df.reset_index()
user_nn_items_df = user_nn_items_df.drop(columns=['level_1'])
user_nn_items_df.rename(columns={'level_0': 'userId', 0: 'movieId'}, inplace=True)
merged_df= pd.merge(train_df,user_nn_items_df,how='inner',on=['userId','movieId'])
#concatenate data for users who rated less than 50 movies merged_df
user_ids_without_nn = set(list(train_df['userId'].unique())) - set(user_ids_nn)
user_ids_without_nn_df = pd.DataFrame(user_ids_without_nn,columns =['userId'])
user_ids_without_nn_df = pd.merge(train_df,user_ids_without_nn_df,how='inner',on='userId')
# ANN reduced matrix
train_concat_df = pd.concat([user_ids_without_nn_df, merged_df])
#train_concat_df = merged_df
return train_concat_df
train_concat_df = def run_ann(user_inner_ids, V, train_df)
def run_ncf(train_concat_df,test_df):
train=train_df[['userId' ,'movieId' ,'rating']]
test=test_df[['userId' ,'movieId' ,'rating']]
train.rename({'userId': 'userID', 'movieId': 'itemID'}, axis=1, inplace=True)
test.rename({'userId': 'userID', 'movieId': 'itemID'}, axis=1, inplace=True)
test=pd.merge(test,pd.DataFrame(np.unique(train['userID']),columns=['userID']),on=['userID'],how='inner')
data = NCFDataset(train=train, test=test, seed=SEED)
EPOCHS = 100
BATCH_SIZE = 256
model = NCF (
n_users=data.n_users,
n_items=data.n_items,
model_type="NeuMF",
n_factors=4,
layer_sizes=[16,8,4],
n_epochs=EPOCHS,
batch_size=BATCH_SIZE,
learning_rate=1e-3,
verbose=10,
seed=SEED)
model.fit(data)
users, items, preds = [], [], []
item = list(train.itemID.unique())
for user in train.userID.unique():
user = [user] * len(item)
users.extend(user)
items.extend(item)
preds.extend(list(model.predict(user, item, is_list=True)))
all_predictions = pd.DataFrame(data={"userID": users, "itemID":items, "prediction":preds})
merged = pd.merge(train, all_predictions, on=["userID", "itemID"], how="outer")
all_predictions = merged[merged.rating.isnull()].drop('rating', axis=1)
return all_predictions
ann_ncf_predictions = run_ncf(train_concat_df, test_df)
return ann_ncf_predictions
pipeline_predictions = run_ann_ncf_pipeline(train, test, user_freq)
```
|
github_jupyter
|
!pip install fastFM
!pip install surprise
!pip install lightfm
!pip install hyperopt
!pip uninstall tensorflow
!pip install tensorflow==1.15
!pip install annoy
import pandas as pd
import math
import numpy as np
import random
from numpy.linalg import inv
from numpy.linalg import multi_dot
import matplotlib.pyplot as plt
import itertools
import warnings
from math import sqrt
from sklearn.feature_extraction import DictVectorizer
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder
from fastFM import als
from sklearn.metrics import mean_squared_error
from lightfm.datasets import fetch_movielens
from lightfm import LightFM
from pydrive.auth import GoogleAuth
from pydrive.drive import GoogleDrive
from google.colab import auth
from oauth2client.client import GoogleCredentials
warnings.filterwarnings("ignore")
from scipy import sparse
from lightfm import LightFM
from sklearn.metrics.pairwise import cosine_similarity
from hyperopt import tpe, fmin, hp, Trials, STATUS_OK,space_eval
import sys
import seaborn as sns
from timeit import Timer
from datetime import datetime
from hyperopt import tpe, fmin, hp, Trials, STATUS_OK,space_eval
from collections import defaultdict
import random
from sklearn.metrics import mean_squared_error
from numpy.linalg import inv
import pandas as pd
import numpy as np
import math
from numpy.linalg import inv
from numpy.linalg import multi_dot
import matplotlib.pyplot as plt
import itertools
import warnings
warnings.filterwarnings("ignore")
from annoy import AnnoyIndex
from ncf_singlenode import *
from dataset import Dataset as NCFDataset
from constants import SEED as DEFAULT_SEED
SEED = DEFAULT_SEED
def get_data():
!pip install -U -q PyDrive
from pydrive.auth import GoogleAuth
from pydrive.drive import GoogleDrive
from google.colab import auth
from oauth2client.client import GoogleCredentials
# Authenticate and create the PyDrive client.
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
id = "1FFYnZRIuzQLeBkDUuJpK_oRtUmdxMd9O"
downloaded = drive.CreateFile({'id':id})
downloaded.GetContentFile('final_dataset.csv')
data = pd.read_csv('final_dataset.csv')
return data
data = get_data()
def train_test_split(data):
user_freq=data.groupby(['userId']).size().reset_index(name='counts')
users_lt3=user_freq[user_freq['counts']<3][['userId']]
users_ge3=user_freq[user_freq['counts']>=3][['userId']]
train1=pd.merge(data, users_lt3, on=['userId'],how='inner')
data1=pd.merge(data, users_ge3, on=['userId'],how='inner')
data1.sort_values(['userId', 'timestamp'], ascending=[True, False],inplace=True)
test=data1.groupby('userId').sample(frac=.3, random_state=2)
test_idx=data1.index.isin(test.index.to_list())
train=train1.append(data1[~test_idx])
return train, test, user_freq
train, test, user_freq = train_test_split(data)
def run_ann_ncf_pipeline(train, test, user_freq):
train_df = train
test_df = test
def create_SVD_UV(train,test,user_freq):
train=train[['userId', 'movieId', 'rating']]
test=test[['userId', 'movieId', 'rating']]
reader = Reader(rating_scale=(1,5))
train = Dataset.load_from_df(train[['userId', 'movieId', 'rating']], reader=reader)
test = Dataset.load_from_df(test[['userId', 'movieId', 'rating']], reader=reader)
raw_ratings = test.raw_ratings
threshold = int(1 * len(raw_ratings))
A_raw_ratings = raw_ratings[:threshold]
test = test.construct_testset(A_raw_ratings)
raw_ratings1 = train.raw_ratings
threshold = int(1 * len(raw_ratings1))
B_raw_ratings = raw_ratings1[:threshold]
train_test = train.construct_testset(B_raw_ratings)
model = SVD(n_epochs=50,n_factors=15,reg_all=0.1,lr_all=0.02)
trainset = train.build_full_trainset()
model.fit(trainset)
# Retrieiving inner ids, as used by surprise package during model training
user_inner_ids = [x for x in trainset.all_users()]
item_inner_ids = [i for i in trainset.all_items()]
# All ids mapped back to values in the actual train set
user_raw_ids = [trainset.to_raw_uid(x) for x in user_inner_ids]
item_raw_ids = [trainset.to_raw_iid(x) for x in item_inner_ids]
U = model.pu
V = model.qi
return U, V, trainset, user_inner_ids
U,V, trainset, user_inner_ids = create_SVD_UV(train,test,user_freq)
def run_ann(user_inner_ids, V, train_df):
#To be used for serialized process; user item search
f = 15 # n_factors
t = AnnoyIndex(f, 'angular') # Length of item vector that will be indexed
for i,v in zip(user_inner_ids,V):
t.add_item(i, v)
t.build(100)
t.save('test_user_item.ann')
u = AnnoyIndex(f, 'angular')
u.load('test_user_item.ann')
# super fast, will just map the file
user_ids_nn = list(user_freq[user_freq['num_movies']>50]['userId'].values)
def find_nn_greater_than_k(user_id, k=100):
return t.get_nns_by_vector(U[trainset.to_inner_uid(user_id)], k)
user_nn_items = list(map(find_nn_greater_than_k,user_ids_nn))
user_nn_items_dict={}
for userid,items in zip(user_ids_nn,user_nn_items):
user_nn_items_dict[userid] = [trainset.to_raw_iid(i) for i in items]
user_nn_items_df = pd.concat({k: pd.Series(v) for k, v in user_nn_items_dict.items()})
user_nn_items_df = user_nn_items_df.reset_index()
user_nn_items_df = user_nn_items_df.drop(columns=['level_1'])
user_nn_items_df.rename(columns={'level_0': 'userId', 0: 'movieId'}, inplace=True)
merged_df= pd.merge(train_df,user_nn_items_df,how='inner',on=['userId','movieId'])
#concatenate data for users who rated less than 50 movies merged_df
user_ids_without_nn = set(list(train_df['userId'].unique())) - set(user_ids_nn)
user_ids_without_nn_df = pd.DataFrame(user_ids_without_nn,columns =['userId'])
user_ids_without_nn_df = pd.merge(train_df,user_ids_without_nn_df,how='inner',on='userId')
# ANN reduced matrix
train_concat_df = pd.concat([user_ids_without_nn_df, merged_df])
#train_concat_df = merged_df
return train_concat_df
train_concat_df = def run_ann(user_inner_ids, V, train_df)
def run_ncf(train_concat_df,test_df):
train=train_df[['userId' ,'movieId' ,'rating']]
test=test_df[['userId' ,'movieId' ,'rating']]
train.rename({'userId': 'userID', 'movieId': 'itemID'}, axis=1, inplace=True)
test.rename({'userId': 'userID', 'movieId': 'itemID'}, axis=1, inplace=True)
test=pd.merge(test,pd.DataFrame(np.unique(train['userID']),columns=['userID']),on=['userID'],how='inner')
data = NCFDataset(train=train, test=test, seed=SEED)
EPOCHS = 100
BATCH_SIZE = 256
model = NCF (
n_users=data.n_users,
n_items=data.n_items,
model_type="NeuMF",
n_factors=4,
layer_sizes=[16,8,4],
n_epochs=EPOCHS,
batch_size=BATCH_SIZE,
learning_rate=1e-3,
verbose=10,
seed=SEED)
model.fit(data)
users, items, preds = [], [], []
item = list(train.itemID.unique())
for user in train.userID.unique():
user = [user] * len(item)
users.extend(user)
items.extend(item)
preds.extend(list(model.predict(user, item, is_list=True)))
all_predictions = pd.DataFrame(data={"userID": users, "itemID":items, "prediction":preds})
merged = pd.merge(train, all_predictions, on=["userID", "itemID"], how="outer")
all_predictions = merged[merged.rating.isnull()].drop('rating', axis=1)
return all_predictions
ann_ncf_predictions = run_ncf(train_concat_df, test_df)
return ann_ncf_predictions
pipeline_predictions = run_ann_ncf_pipeline(train, test, user_freq)
| 0.361728 | 0.420659 |
# 变分量子本征求解器
<em> Copyright (c) 2021 Institute for Quantum Computing, Baidu Inc. All Rights Reserved. </em>
## 概览
目前普遍认为,量子计算在近期很有前景的一个应用是处理量子化学问题 [1-2]。**变分量子本征求解器** (VQE)作为这个研究方向的核心应用之一,为研究者们提供了可以在目前含噪的中等规模量子设备(NISQ device)上研究量子化学的可能 [1-4]。其核心任务是求解一个量子尺度上封闭物理系统的哈密顿量 $\hat{H}$ 的基态能量及其对应的量子态。主要的实现方法是通过在量子设备上准备一个参数化的试探波函数 $|\Psi(\boldsymbol\theta)\rangle$ 然后结合经典机器学习中的优化算法(例如梯度下降法)去不断地调整、优化参数 $\boldsymbol\theta$ 使得期望值 $\langle \Psi(\boldsymbol\theta)|\hat{H}|\Psi(\boldsymbol\theta)\rangle$ 最小化。这套方案的基本原理是基于 **Rayleigh-Ritz 变分原理**。
$$
E_0 = \min_{\boldsymbol\theta} \langle \Psi(\boldsymbol\theta)|\hat{H}|\Psi(\boldsymbol\theta)\rangle.
\tag{1}
$$
其中 $E_0$ 表示该系统的基态能量。从数值分析的角度来看,该问题可以被理解为求解一个**离散化**哈密顿量 $H$(埃尔米特矩阵)的最小本征值 $\lambda_{\min}$ 和其对应的本征向量 $|\Psi_0\rangle$。具体的离散化过程是如何通过建立模型实现的,这属于量子化学的专业领域范畴。精确地解释该过程需要很长的篇幅,这超过了本教程所能处理的范围。我们会在下一节背景知识模块粗略的介绍一下相关知识,感兴趣的读者可以参考 `量子化学: 基本原理和从头计算法`系列丛书 [5]。通常来说,为了能在量子设备上处理量子化学问题,哈密顿量 $H$ 会被表示成为泡利算符 $\{X,Y,Z\}$ 的加权求和形式。
$$
H = \sum_k c_k ~ \bigg( \bigotimes_{j=0}^{M-1} \sigma_j^{(k)} \bigg),
\tag{2}
$$
其中 $c_k$ 表示权重系数, $\sigma_j^{(k)} \in \{I,X,Y,Z\}$ 并且 $M$ 表示所需的量子比特个数。这样一种哈密顿量的表示形式被称为 **泡利字符串**。以下为一个2量子比特的具体例子,
$$
H= 0.12~Y_0 \otimes I_1-0.04~X_0\otimes Z_1.
\tag{3}
$$
在下一节,我们会补充一些关于电子结构问题的背景知识。本质上讨论的就是上述哈密顿量 $H$ 是如何计算的。对于熟悉相关背景的读者,或者主要关心如何在量桨上实现 VQE 的读者,请直接跳转至第三节分析氢分子($H_2$)基态的具体例子。
## 背景: 电子结构问题
这里,我们集中讨论下量子化学中的一个基本问题 -- **电子结构问题**。更准确的说,我们关心的是给定分子(molecule)的低位能量本征态。这些信息可以帮助我们预测化学反应的速率和分子的稳定结构等等 [6]。假设一个分子由 $N_n$ 个原子核和 $N_e$ 个电子组成,描述该分子系统总能量的哈密顿量算符 $\hat{H}_{mol}$ 在一次量子化表示下可以写为,
$$
\begin{align}
\hat{H}_{\text{mol}} & = -\sum_{i}\frac{\nabla_{R_i}^2}{2M_i} - \sum_{i} \frac{\nabla_{r_i}^2}{2} -\sum_{i,j}\frac{Z_i}{\lvert R_i - r_j\lvert} + \sum_{i,j>i}\frac{Z_iZ_j}{\lvert R_i - R_j\lvert} + \sum_{i, j>i}\frac{1}{\lvert r_i - r_j\lvert},
\tag{4}
\end{align}
$$
其中 $R_i、M_i$ 和 $Z_i$ 分别表示第 $i$ 个原子核的位置、质量和原子序数(原子核内质子数),第 $i$ 个电子的位置则表示为 $r_i$。以上公式右边前两项分别代表原子核和电子的总动能。第三项表示带正电的质子和带负电的电子之间的库伦相互吸引作用。最后两项则表示原子核-原子核之间,电子-电子之间的相互排斥作用。这里,分子哈密顿量 $\hat{H}_\text{mol}$ 使用的是原子单位制能量 **哈特里能量**(Hartree),记为 Ha。$1$ 哈特里能量的大小为 $[\hbar^2/(m_ee^2a_0^2)] = 27.2$ 电子伏或 $630$ 千卡/摩尔,其中 $m_e、e$ 和 $a_0$ 分别表示电子质量、基本电荷和玻尔半径。
**注释1:** 在处理电子结构问题时,我们不考虑自旋-轨道耦合以及超精细结构。如果出于计算需要,可以作为微扰加入。
### 玻恩-奥本海默近似
由于原子核的质量要远大于电子,因而在同样的相互作用下电子的运动速度会比原子核快很多。所以,将原子核所处的位置看成固定 $R_i =$常数 是一种合理的近似。这种通过在时间尺度上将电子行为和原子核行为去耦合的近似处理思想被称为玻恩-奥本海默近似。作为近似的直接结果,公式(4)中原子核的动能项会被消去并且表示原子核-原子核相互排斥作用的项可以被认为是一个能量移位(这个项是与电子位置 $r_i$ 无关的)从而也可以作为常数项被忽略。经过这些步骤后,我们可以把哈密顿量近似为:
$$
\begin{align}
\hat{H}_{\text{electron}} & = - \sum_{i} \frac{\nabla_{r_i}^2}{2} -\sum_{i,j}\frac{Z_i}{\lvert R_i - r_j\lvert} + \sum_{i, j>i}\frac{1}{\lvert r_i - r_j\lvert}
\tag{5},
\end{align}
$$
在经过以上近似后,分子中多电子结构的能级在理论上可以通过求解以下不含时薛定谔方程获得:
$$
\hat{H}_{\text{electron}} |\Psi_n \rangle = E_n |\Psi_n \rangle,
\tag{6}
$$
其中 $n$ 指代能级。值得注意的是,电子哈密顿量中电子-电子相互排斥作用的求和项数会随着电子数 $N_e$ 的增多至 $N_e(N_e-1)/2$ 项。这意味着对于一个含有16个电子的氧分子($O_2$)我们需要计算多达120项的相互排斥作用项。 一般来说,这样的问题是无法从理论上精确求解的。正如狄拉克在 [Quantum mechanics of many-electron systems](https://royalsocietypublishing.org/doi/10.1098/rspa.1929.0094) [7] 所指出的那样,
> *The underlying physical laws necessary for the mathematical theory of a large part of physics and the whole of chemistry are thus completely known, and the difficulty is only that the exact application of these laws leads to equations much too complicated to be soluble.*
>
> -- Paul Dirac (1929)
由于解析的方法太复杂,那么我们可以采用数值方法来处理。一个最简单的数值方法(离散化方法)就是把上述作用中无限维度希尔伯特空间离散化为等间距排开的立方体晶格点。在这样一个离散化的空间里,主要运算规则为复数域的线性代数。假设空间的每个轴都离散为等间距排开的 $k$ 个点,则 $N$-电子(为了方便去掉下标 $e$)的多体波函数可以写为 [2]:
$$
|\Psi \rangle = \sum_{\mathbf{x_1}, \ldots, \mathbf{x_N}} \psi(\mathbf{x_1}, \ldots, \mathbf{x_N}) \mathcal{A}(|\mathbf{x_1}, \ldots, \mathbf{x_N}\rangle).
\tag{7}
$$
其中坐标 $|\mathbf{x_j}\rangle = |r_j\rangle |\sigma_j\rangle$ 记录第 $j$ 个电子的空间位置信息和自旋,$|r_j\rangle = |x_j,y_j,z_j\rangle$ 且 $j\in \{1,2,\cdots,N\}$, $x_j,y_j,z_j \in \{0,1,\cdots,k-1\}$ 同时 $\sigma_j \in \{\downarrow,\uparrow\}$ 表示自旋向下和向上。这样一种离散化方式共计需要 $k^{3N}\times 2^{N}$ 个数据来表示波函数。在这里,$\mathcal{A}$ 表示反对称化操作(根据泡利不相容原理)并且 $\psi(\mathbf{x_1}, \mathbf{x_2}, \ldots, \mathbf{x_N})=\langle\mathbf{x_1}, \mathbf{x_2}, \ldots, \mathbf{x_N}|\Psi\rangle$。 可以看出,经典计算机存储这样一个波函数需要的内存是随着电子个数呈指数增长的。这使得基于这种离散化的经典数值方法,无法模拟超过几十个电子的系统。那么,我们是不是能够通过量子设备来存储和准备这样一个波函数然后求解基态能量 $E_0$ 呢?在下一节中,我们将以最简单的分子系统 -- 氢分子($H_2$)为例,讲解 VQE 算法。
**注释2:** 关于量子化学和现有数值计算方法的综述也超过了本教程的处理范围,我们推荐感兴趣的读者去查阅以下经典教材 Helgaker 等人撰写的 *'Molecular Electronic-Structure Theory'* [6] 以及 Szabo & Ostlund 撰写的 *'Modern Quantum Chemistry: Introduction to Advanced Electronic Structure Theory'* [8]。 如果需要弥补量子计算和量子化学之间知识空缺,请参考以下综述文章 [Quantum chemistry in the age of quantum computing](https://pubs.acs.org/doi/10.1021/acs.chemrev.8b00803) [1] 和 [Quantum computational chemistry](https://journals.aps.org/rmp/abstract/10.1103/RevModPhys.92.015003) [2] 。
**注释3:** 对于量子化学中的能量计算,我们期望能够达到 **化学精度**(chemical accuracy)$1.6\times10^{-3}$ Ha 或者 1 千卡/摩尔。
## 氢分子 $H_2$ 基态能量
### 构造电子哈密顿量
首先,让我们通过下面几行代码引入必要的 library 和 package。量桨的量子化学工具包是基于 `psi4` 和 `openfermion` 进行开发的,所以需要读者先行安装这两个语言包。在进入下面的教程之前,我们强烈建议您先阅读[哈密顿量的构造](./BuildingMolecule_CN.ipynb)教程,该教程介绍了如何使用量桨的量子化学工具包。
**注意:关于环境设置,请参考 [README_CN.md](https://github.com/PaddlePaddle/Quantum/blob/master/README_CN.md).**
```
import paddle
import paddle_quantum.qchem as qchem
from paddle_quantum.utils import Hamiltonian
from paddle_quantum.circuit import UAnsatz
import os
import matplotlib.pyplot as plt
import numpy
from numpy import pi as PI
from numpy import savez, zeros
# 无视警告
import warnings
warnings.filterwarnings("ignore")
```
对于具体需要分析的分子,我们需要其**几何构型** (geometry)、**基组**(basis set,例如 STO-3G 基于高斯函数)、**多重度**(multiplicity)以及**分子的净电荷数** (charge) 等多项信息来建模计算出该分子单体积分 (one-body integrations),双体积分(two-body integrations) 以及哈密顿量等信息。接下来,通过量桨的量子化学工具包将分子的哈密顿量提取出来并储存为 paddle quantum 的 `Hamiltonian` 类,方便我们下一步的操作。
```
geo = qchem.geometry(structure=[['H', [-0., 0., 0.0]], ['H', [-0., 0., 0.74]]])
# geo = qchem.geometry(file='h2.xyz')
# 将分子信息存储在 molecule 里,包括单体积分(one-body integrations),双体积分(two-body integrations),分子的哈密顿量等
molecule = qchem.get_molecular_data(
geometry=geo,
basis='sto-3g',
charge=0,
multiplicity=1,
method="fci",
if_save=True,
if_print=True
)
# 提取哈密顿量
molecular_hamiltonian = qchem.spin_hamiltonian(molecule=molecule,
filename=None,
multiplicity=1,
mapping_method = 'jordan_wigner',)
# 打印结果
print("\nThe generated h2 Hamiltonian is \n", molecular_hamiltonian)
```
**注释4:** 生成这个哈密顿量的几何构型中,两个氢原子间的原子间隔(interatomic distance)为 $d = 74$ pm。
除了输入分子的几何结构外,我们还支持读取分子的几何构型文件 (`.xyz` 文件),关于量子化学工具包更多的用法请参考[哈密顿量的构造](./BuildingMolecule_CN.ipynb)教程。如果你需要测试更多分子的几何构型,请移步至这个[数据库](http://smart.sns.it/molecules/index.html)。
### 搭建量子神经网络(QNN)和试探波函数
在实现VQE的过程中,我们首先需要设计量子神经网络QNN(也可以理解为参数化量子电路)来准备试探波函数 $|\Psi(\boldsymbol\theta)\rangle$。这里,我们提供一个预设好的的深度为 $D$ 层的 4-量子比特的量子电路模板,图中的虚线框内为一层:

- 我们预设一些该参数化电路的参数,比如宽度为 $N = 4$ 量子位。
- 初始化其中的变量参数,${\bf{\theta }}$ 代表我们量子神经网络中的参数组成的向量。
接下来我们根据上图中的电路设计,通过 Paddle Quantum 的 `UAnsatz` 函数和内置的 `real_entangled_layer(theta, D)` 电路模板来高效搭建量子神经网络。
```
def U_theta(theta, Hamiltonian, N, D):
"""
Quantum Neural Network
"""
# 按照量子比特数量/网络宽度初始化量子神经网络
cir = UAnsatz(N)
# 内置的 {R_y + CNOT} 电路模板
cir.real_entangled_layer(theta[:D], D)
# 铺上最后一列 R_y 旋转门
for i in range(N):
cir.ry(theta=theta[D][i][0], which_qubit=i)
# 量子神经网络作用在默认的初始态 |0000> 上
fin_state = cir.run_state_vector()
# 计算给定哈密顿量的期望值
expectation_val = cir.expecval(Hamiltonian)
return expectation_val, cir, fin_state
```
### 配置训练模型 - 损失函数
现在我们已经有了数据和量子神经网络的架构,我们将进一步定义训练参数、模型和损失函数。通过作用量子神经网络 $U(\theta)$ 在初始态 $|0..0\rangle$ 上,我们将得到输出态 $\left| {\psi \left( {\bf{\theta }} \right)} \right\rangle $。进一步,在VQE模型中的损失函数一般由量子态 $\left| {\psi \left( {\bf{\theta }} \right)} \right\rangle$ 关于哈密顿量 $H$ 的期望值 (能量期望值 expectation value) 给出,
$$
\min_{\boldsymbol\theta} \mathcal{L}(\boldsymbol \theta) = \min_{\boldsymbol\theta} \langle \Psi(\boldsymbol\theta)|H |\Psi(\boldsymbol\theta)\rangle
= \min_{\boldsymbol\theta} \sum_k c_k~\langle \Psi(\boldsymbol\theta)| \bigotimes_j \sigma_j^{(k)}|\Psi(\boldsymbol\theta)\rangle.
\tag{8}
$$
```
class StateNet(paddle.nn.Layer):
"""
Construct the model net
"""
def __init__(self, shape, dtype="float64"):
super(StateNet, self).__init__()
# 初始化 theta 参数列表,并用 [0, 2*pi] 的均匀分布来填充初始值
self.theta = self.create_parameter(shape=shape,
default_initializer=paddle.nn.initializer.Uniform(low=0.0, high=2*PI),
dtype=dtype, is_bias=False)
# 定义损失函数和前向传播机制
def forward(self, N, D):
# 计算损失函数/期望值
loss, cir, fin_state = U_theta(self.theta, molecular_hamiltonian.pauli_str, N, D)
return loss, cir, fin_state
```
### 配置训练模型 - 模型参数
在进行量子神经网络的训练之前,我们还需要进行一些训练的超参数设置,主要是学习速率(LR, learning rate)、迭代次数(ITR, iteration)和量子神经网络计算模块的深度(D, Depth)。这里我们设定学习速率为 0.5, 迭代次数为 50 次。读者不妨自行调整来直观感受下超参数调整对训练效果的影响。
```
ITR = 80 # 设置训练的总迭代次数
LR = 0.4 # 设置学习速率
D = 2 # 设置量子神经网络中重复计算模块的深度 Depth
N = molecular_hamiltonian.n_qubits # 设置参与计算的量子比特数
```
### 进行训练
当训练模型的各项参数都设置完成后,我们将数据转化为 Paddle 中的张量,进而进行量子神经网络的训练。过程中我们用的是Adam Optimizer,也可以调用Paddle中提供的其他优化器。我们将训练过程中的结果存储在summary_data文件中。
```
# 确定网络的参数维度
net = StateNet(shape=[D + 1, N, 1])
# 一般来说,我们利用Adam优化器来获得相对好的收敛,
# 当然你可以改成SGD或者是RMS prop.
opt = paddle.optimizer.Adam(learning_rate=LR, parameters=net.parameters())
# 记录优化结果
summary_iter, summary_loss = [], []
# 优化循环
for itr in range(1, ITR + 1):
# 前向传播计算损失函数
loss, cir, fin_state = net(N, D)
# 在动态图机制下,反向传播极小化损失函数
loss.backward()
opt.minimize(loss)
opt.clear_grad()
# 更新优化结果
summary_loss.append(loss.numpy())
summary_iter.append(itr)
# 打印结果
if itr % 20 == 0:
print("iter:", itr, "loss:", "%.4f" % loss.numpy())
print("iter:", itr, "Ground state energy:", "%.4f Ha"
% loss.numpy())
if itr == ITR:
print("\n训练后的电路:")
print(cir)
# 储存训练结果到 output 文件夹
os.makedirs("output", exist_ok=True)
savez("./output/summary_data", iter = summary_iter,
energy=summary_loss)
```
### 测试效果
我们现在已经完成了量子神经网络的训练,通过 VQE 得到的基态能量的估计值大致为 $E_0 \approx -1.137$ Ha,这与通过全价构型相互作用(FCI)$E_0 = -1.13728$ Ha 计算得出的值是在化学精度 $\varepsilon = 1.6 \times 10^{-3}$ Ha 内相符合的。
```
result = numpy.load('./output/summary_data.npz')
eig_val, eig_state = numpy.linalg.eig(
molecular_hamiltonian.construct_h_matrix())
min_eig_H = numpy.min(eig_val.real)
min_loss = numpy.ones([len(result['iter'])]) * min_eig_H
plt.figure(1)
func1, = plt.plot(result['iter'], result['energy'],
alpha=0.7, marker='', linestyle="-", color='r')
func_min, = plt.plot(result['iter'], min_loss,
alpha=0.7, marker='', linestyle=":", color='b')
plt.xlabel('Number of iteration')
plt.ylabel('Energy (Ha)')
plt.legend(handles=[
func1,
func_min
],
labels=[
r'$\left\langle {\psi \left( {\theta } \right)} '
r'\right|H\left| {\psi \left( {\theta } \right)} \right\rangle $',
'Ground-state energy',
], loc='best')
#plt.savefig("vqe.png", bbox_inches='tight', dpi=300)
plt.show()
```
## 通过 VQE 确定原子间隔
还记得在前面的注释中提到我们默认使用的两个氢原子间原子间隔为 $74$ pm 吗?VQE 的另一个用法便是通过在不同的原子间隔下多次运行然后观察运行结果的最小值是在什么原子间隔发生的,这个间隔即为估计得真实原子间隔。

从上图可以看出,最小值确实发生在 $d = 74$ pm (1 pm = $1\times 10^{-12}$m) 附近,这是与[实验测得数据](https://cccbdb.nist.gov/exp2x.asp?casno=1333740&charge=0)相符合的 $d_{exp} (H_2) = 74.14$ pm.
_______
## 参考文献
[1] Cao, Yudong, et al. Quantum Chemistry in the Age of Quantum Computing. [Chemical reviews 119.19 (2019): 10856-10915.](https://pubs.acs.org/doi/10.1021/acs.chemrev.8b00803)
[2] McArdle, Sam, et al. Quantum computational chemistry. [Reviews of Modern Physics 92.1 (2020): 015003.](https://journals.aps.org/rmp/abstract/10.1103/RevModPhys.92.015003)
[3] Peruzzo, A. et al. A variational eigenvalue solver on a photonic quantum processor. [Nat. Commun. 5, 4213 (2014).](https://www.nature.com/articles/ncomms5213)
[4] Moll, Nikolaj, et al. Quantum optimization using variational algorithms on near-term quantum devices. [Quantum Science and Technology 3.3 (2018): 030503.](https://iopscience.iop.org/article/10.1088/2058-9565/aab822)
[5] 徐光宪, 黎乐民, 王德民. 量子化学: 基本原理和从头计算法(上)[M], 第二版. 北京: 科学出版社, 2012;
[6] Helgaker, Trygve, Poul Jorgensen, and Jeppe Olsen. Molecular electronic-structure theory. John Wiley & Sons, 2014.
[7] Dirac, Paul Adrien Maurice. Quantum mechanics of many-electron systems. [Proceedings of the Royal Society of London. Series A, Containing Papers of a Mathematical and Physical Character 123.792 (1929): 714-733.](https://royalsocietypublishing.org/doi/10.1098/rspa.1929.0094)
[8] Szabo, Attila, and Neil S. Ostlund. Modern quantum chemistry: introduction to advanced electronic structure theory. Courier Corporation, 2012.
|
github_jupyter
|
import paddle
import paddle_quantum.qchem as qchem
from paddle_quantum.utils import Hamiltonian
from paddle_quantum.circuit import UAnsatz
import os
import matplotlib.pyplot as plt
import numpy
from numpy import pi as PI
from numpy import savez, zeros
# 无视警告
import warnings
warnings.filterwarnings("ignore")
geo = qchem.geometry(structure=[['H', [-0., 0., 0.0]], ['H', [-0., 0., 0.74]]])
# geo = qchem.geometry(file='h2.xyz')
# 将分子信息存储在 molecule 里,包括单体积分(one-body integrations),双体积分(two-body integrations),分子的哈密顿量等
molecule = qchem.get_molecular_data(
geometry=geo,
basis='sto-3g',
charge=0,
multiplicity=1,
method="fci",
if_save=True,
if_print=True
)
# 提取哈密顿量
molecular_hamiltonian = qchem.spin_hamiltonian(molecule=molecule,
filename=None,
multiplicity=1,
mapping_method = 'jordan_wigner',)
# 打印结果
print("\nThe generated h2 Hamiltonian is \n", molecular_hamiltonian)
def U_theta(theta, Hamiltonian, N, D):
"""
Quantum Neural Network
"""
# 按照量子比特数量/网络宽度初始化量子神经网络
cir = UAnsatz(N)
# 内置的 {R_y + CNOT} 电路模板
cir.real_entangled_layer(theta[:D], D)
# 铺上最后一列 R_y 旋转门
for i in range(N):
cir.ry(theta=theta[D][i][0], which_qubit=i)
# 量子神经网络作用在默认的初始态 |0000> 上
fin_state = cir.run_state_vector()
# 计算给定哈密顿量的期望值
expectation_val = cir.expecval(Hamiltonian)
return expectation_val, cir, fin_state
class StateNet(paddle.nn.Layer):
"""
Construct the model net
"""
def __init__(self, shape, dtype="float64"):
super(StateNet, self).__init__()
# 初始化 theta 参数列表,并用 [0, 2*pi] 的均匀分布来填充初始值
self.theta = self.create_parameter(shape=shape,
default_initializer=paddle.nn.initializer.Uniform(low=0.0, high=2*PI),
dtype=dtype, is_bias=False)
# 定义损失函数和前向传播机制
def forward(self, N, D):
# 计算损失函数/期望值
loss, cir, fin_state = U_theta(self.theta, molecular_hamiltonian.pauli_str, N, D)
return loss, cir, fin_state
ITR = 80 # 设置训练的总迭代次数
LR = 0.4 # 设置学习速率
D = 2 # 设置量子神经网络中重复计算模块的深度 Depth
N = molecular_hamiltonian.n_qubits # 设置参与计算的量子比特数
# 确定网络的参数维度
net = StateNet(shape=[D + 1, N, 1])
# 一般来说,我们利用Adam优化器来获得相对好的收敛,
# 当然你可以改成SGD或者是RMS prop.
opt = paddle.optimizer.Adam(learning_rate=LR, parameters=net.parameters())
# 记录优化结果
summary_iter, summary_loss = [], []
# 优化循环
for itr in range(1, ITR + 1):
# 前向传播计算损失函数
loss, cir, fin_state = net(N, D)
# 在动态图机制下,反向传播极小化损失函数
loss.backward()
opt.minimize(loss)
opt.clear_grad()
# 更新优化结果
summary_loss.append(loss.numpy())
summary_iter.append(itr)
# 打印结果
if itr % 20 == 0:
print("iter:", itr, "loss:", "%.4f" % loss.numpy())
print("iter:", itr, "Ground state energy:", "%.4f Ha"
% loss.numpy())
if itr == ITR:
print("\n训练后的电路:")
print(cir)
# 储存训练结果到 output 文件夹
os.makedirs("output", exist_ok=True)
savez("./output/summary_data", iter = summary_iter,
energy=summary_loss)
result = numpy.load('./output/summary_data.npz')
eig_val, eig_state = numpy.linalg.eig(
molecular_hamiltonian.construct_h_matrix())
min_eig_H = numpy.min(eig_val.real)
min_loss = numpy.ones([len(result['iter'])]) * min_eig_H
plt.figure(1)
func1, = plt.plot(result['iter'], result['energy'],
alpha=0.7, marker='', linestyle="-", color='r')
func_min, = plt.plot(result['iter'], min_loss,
alpha=0.7, marker='', linestyle=":", color='b')
plt.xlabel('Number of iteration')
plt.ylabel('Energy (Ha)')
plt.legend(handles=[
func1,
func_min
],
labels=[
r'$\left\langle {\psi \left( {\theta } \right)} '
r'\right|H\left| {\psi \left( {\theta } \right)} \right\rangle $',
'Ground-state energy',
], loc='best')
#plt.savefig("vqe.png", bbox_inches='tight', dpi=300)
plt.show()
| 0.489748 | 0.944022 |
```
import os
import glob
import time
import numpy as np
from pyarrow import fs
import pyarrow.parquet as pq
from sklearn.neighbors import BallTree
is_index_file_exist = os.path.isfile("../../../data/Master/New/index.parquet")
files = glob.glob(os.path.join('../../../','data','Master', 'New', '*[0-9].parquet')) if is_index_file_exist else []
## Interested Dimensions in the GNAF Files
interested_dims = ['LATITUDE', 'LONGITUDE', 'FULL_ADDRESS', 'STATE', 'SA4_NAME_2016',
'LGA_NAME_2016', 'SSC_NAME_2016', 'SA3_NAME_2016', 'SA2_NAME_2016', 'ADDRESS_DETAIL_PID']
local = fs.LocalFileSystem()
# Set Minimum and Maximum lat for all properties within Australia
lat_min = -43.58301104
lat_max = -9.23000371
lon_min = 96.82159219
lon_max = 167.99384663
# 1 lat equals 110.574km
deg = 110.574
# Conversion Rate - radians to kilometer
rad_to_km = 6371
def load_parquet(lat, lon, distance):
df = pq.read_table(
files,
filesystem = local,
columns = interested_dims,
filters=
[('LATITUDE', '>=', lat - distance),
('LATITUDE', '<=', lat + distance),
('LONGITUDE', '>=', lon - distance),
('LONGITUDE', '<=', lon + distance)
]).\
to_pandas()
return df
def ensure_lat_lon_within_range(lat, lon):
# Ensure Latitudge within the AU range
lat = max(lat, lat_min)
lat = min(lat, lat_max)
# Ensure longitutde within the AU range
lon = max(lon, lon_min)
lon = min(lon, lon_max)
return lat,lon
def filter_for_rows_within_mid_distance(df, lat, lon, mid_distance):
mid_df = df[df.LATITUDE.between(lat - middle_distance, lat + middle_distance) &
df.LONGITUDE.between(lon - middle_distance, lon + middle_distance)]
return mid_df
def find_nearest_address(lat, lon, km = None , n = 1):
## 1. Initial distance setting according to lat/lon arguments
lat , lon = ensure_lat_lon_within_range(lat, lon)
min_distance = 0
distance = (km if km else 1) / deg
## 2. Make the first load of GNAF dataset
gnaf_df = load_parquet(lat, lon, distance)
# 2.a If the desired count of addresses not exist, increase the radius
while (gnaf_df.shape[0] < n):
min_distance = distance
distance *= 2
gnaf_df = load_parquet(lat, lon, distance)
print("gnaf_df.shape: First Load: ",gnaf_df.shape)
# 2.b Keep reducing the size of rows if more than 10k adddresses are found within the radius
# Take the median distance to reduce
# This is to limit the number of datapoint to build the Ball tree in the next step
while (gnaf_df.shape[0] >= n + 10000):
middle_distance = (distance - min_distance)/2
gnaf_df = filter_for_rows_within_mid_distance(gnaf_df, lat, lon, middle_distance)
print("gnaf_df.shape: Reduced Load: ", gnaf_df.shape)
distance = middle_distance
print("gnaf_df.shape: Final Load: ", gnaf_df.shape)
## 3. Build the Ball Tree and Query for the nearest within k distance
ball_tree = BallTree(np.deg2rad(gnaf_df[['LATITUDE', 'LONGITUDE']].values), metric='haversine')
distances, indices = ball_tree.query(np.deg2rad(np.c_[lat, lon]), k= min(n, gnaf_df.shape[0]))
# Get indices of the search result, Extract pid and calculate distance(km)
indices = indices[0].tolist()
pids = gnaf_df.ADDRESS_DETAIL_PID.iloc[indices].tolist()
distance_map = dict(zip(pids ,[distance * rad_to_km for distance in distances[0]]))
## 4. Filter the GNAF dataset by address_detail_pid and Extract the interested columns
bool_list = gnaf_df['ADDRESS_DETAIL_PID'].isin(pids)
final_gnaf_df = gnaf_df[bool_list]
final_gnaf_df = final_gnaf_df[interested_dims]
final_gnaf_df['DISTANCE'] = final_gnaf_df['ADDRESS_DETAIL_PID'].map(distance_map)
return final_gnaf_df.sort_values('DISTANCE')
find_nearest_address(-33.3643823,150.1687078, n= 2)
# find_nearest_address(-33.031745, 151.135357, n= 2)
```
|
github_jupyter
|
import os
import glob
import time
import numpy as np
from pyarrow import fs
import pyarrow.parquet as pq
from sklearn.neighbors import BallTree
is_index_file_exist = os.path.isfile("../../../data/Master/New/index.parquet")
files = glob.glob(os.path.join('../../../','data','Master', 'New', '*[0-9].parquet')) if is_index_file_exist else []
## Interested Dimensions in the GNAF Files
interested_dims = ['LATITUDE', 'LONGITUDE', 'FULL_ADDRESS', 'STATE', 'SA4_NAME_2016',
'LGA_NAME_2016', 'SSC_NAME_2016', 'SA3_NAME_2016', 'SA2_NAME_2016', 'ADDRESS_DETAIL_PID']
local = fs.LocalFileSystem()
# Set Minimum and Maximum lat for all properties within Australia
lat_min = -43.58301104
lat_max = -9.23000371
lon_min = 96.82159219
lon_max = 167.99384663
# 1 lat equals 110.574km
deg = 110.574
# Conversion Rate - radians to kilometer
rad_to_km = 6371
def load_parquet(lat, lon, distance):
df = pq.read_table(
files,
filesystem = local,
columns = interested_dims,
filters=
[('LATITUDE', '>=', lat - distance),
('LATITUDE', '<=', lat + distance),
('LONGITUDE', '>=', lon - distance),
('LONGITUDE', '<=', lon + distance)
]).\
to_pandas()
return df
def ensure_lat_lon_within_range(lat, lon):
# Ensure Latitudge within the AU range
lat = max(lat, lat_min)
lat = min(lat, lat_max)
# Ensure longitutde within the AU range
lon = max(lon, lon_min)
lon = min(lon, lon_max)
return lat,lon
def filter_for_rows_within_mid_distance(df, lat, lon, mid_distance):
mid_df = df[df.LATITUDE.between(lat - middle_distance, lat + middle_distance) &
df.LONGITUDE.between(lon - middle_distance, lon + middle_distance)]
return mid_df
def find_nearest_address(lat, lon, km = None , n = 1):
## 1. Initial distance setting according to lat/lon arguments
lat , lon = ensure_lat_lon_within_range(lat, lon)
min_distance = 0
distance = (km if km else 1) / deg
## 2. Make the first load of GNAF dataset
gnaf_df = load_parquet(lat, lon, distance)
# 2.a If the desired count of addresses not exist, increase the radius
while (gnaf_df.shape[0] < n):
min_distance = distance
distance *= 2
gnaf_df = load_parquet(lat, lon, distance)
print("gnaf_df.shape: First Load: ",gnaf_df.shape)
# 2.b Keep reducing the size of rows if more than 10k adddresses are found within the radius
# Take the median distance to reduce
# This is to limit the number of datapoint to build the Ball tree in the next step
while (gnaf_df.shape[0] >= n + 10000):
middle_distance = (distance - min_distance)/2
gnaf_df = filter_for_rows_within_mid_distance(gnaf_df, lat, lon, middle_distance)
print("gnaf_df.shape: Reduced Load: ", gnaf_df.shape)
distance = middle_distance
print("gnaf_df.shape: Final Load: ", gnaf_df.shape)
## 3. Build the Ball Tree and Query for the nearest within k distance
ball_tree = BallTree(np.deg2rad(gnaf_df[['LATITUDE', 'LONGITUDE']].values), metric='haversine')
distances, indices = ball_tree.query(np.deg2rad(np.c_[lat, lon]), k= min(n, gnaf_df.shape[0]))
# Get indices of the search result, Extract pid and calculate distance(km)
indices = indices[0].tolist()
pids = gnaf_df.ADDRESS_DETAIL_PID.iloc[indices].tolist()
distance_map = dict(zip(pids ,[distance * rad_to_km for distance in distances[0]]))
## 4. Filter the GNAF dataset by address_detail_pid and Extract the interested columns
bool_list = gnaf_df['ADDRESS_DETAIL_PID'].isin(pids)
final_gnaf_df = gnaf_df[bool_list]
final_gnaf_df = final_gnaf_df[interested_dims]
final_gnaf_df['DISTANCE'] = final_gnaf_df['ADDRESS_DETAIL_PID'].map(distance_map)
return final_gnaf_df.sort_values('DISTANCE')
find_nearest_address(-33.3643823,150.1687078, n= 2)
# find_nearest_address(-33.031745, 151.135357, n= 2)
| 0.628407 | 0.398377 |
<a href="https://colab.research.google.com/github/Neuralwood-Net/face-recognizer-9000/blob/main/notebooks/pretraining_celeba_cnn_v2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Train images of authors, 64x64px
## Ok network
```
gpu_info = !nvidia-smi
gpu_info = '\n'.join(gpu_info)
if gpu_info.find('failed') >= 0:
print('Select the Runtime > "Change runtime type" menu to enable a GPU accelerator, ')
print('and then re-execute this cell.')
else:
print(gpu_info)
from psutil import virtual_memory
ram_gb = virtual_memory().total / 1e9
print('Your runtime has {:.1f} gigabytes of available RAM\n'.format(ram_gb))
if ram_gb < 20:
print('To enable a high-RAM runtime, select the Runtime > "Change runtime type"')
print('menu, and then select High-RAM in the Runtime shape dropdown. Then, ')
print('re-execute this cell.')
else:
print('You are using a high-RAM runtime!')
```
### Imports
```
import time
import os
import copy
import numpy as np
import matplotlib.pyplot as plt
from tqdm import tqdm
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.utils.data
from torch.optim import lr_scheduler
import torchvision
from torchvision import datasets, models, transforms
from google.cloud import storage
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
device
```
### Read and prepare the data
```
from google.cloud import storage
client = storage.Client()
bucket_name = "tdt4173-datasets"
bucket = client.get_bucket(bucket_name)
zipfilename = "/home/jupyter/data/faces/balanced_sampled_128px_color_130480_images_33_percent_val.zip"
blob_name = "faces/balanced_sampled_128px_color_130480_images_33_percent_val.zip"
blob = bucket.get_blob(blob_name)
blob.download_to_filename(zipfilename)
! rm -rf /home/jupyter/data/faces/images/val/.ipynb_checkpoints
!unzip /home/jupyter/data/faces/images_final_balanced_128px_color_26180_train_ca_300_val_per_class.zip -d /home/jupyter/data/faces/
BATCH_SIZE = 16
data_transforms = transforms.Compose([
transforms.Resize(224),
# transforms.CenterCrop(128),
transforms.Grayscale(num_output_channels=3),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225]),
# transforms.Normalize([0.5], [0.5]),
])
data_dir = '/home/jupyter/data/faces/images'
image_datasets = {x: datasets.ImageFolder(os.path.join(data_dir, x),
data_transforms)
for x in ['train', 'val']}
dataloaders = {x: torch.utils.data.DataLoader(image_datasets[x], batch_size=BATCH_SIZE,
shuffle=True, num_workers=4)
for x in ['train', 'val']}
dataset_sizes = {x: len(image_datasets[x]) for x in ['train', 'val']}
class_names = image_datasets['train'].classes
class_names
image_datasets['val'].classes
dataset_sizes
def imshow(inp, title=None):
"""Imshow for Tensor."""
inp = inp.numpy().transpose((1, 2, 0))
# mean = np.array([0.485, 0.456, 0.406])
# std = np.array([0.229, 0.224, 0.225])
mean = np.array([0.5])
std = np.array([0.5])
inp = std * inp + mean
inp = np.clip(inp, 0, 1)
plt.imshow(inp)
if title is not None:
plt.title(title)
plt.pause(0.001) # pause a bit so that plots are updated
# Get a batch of training data
inputs, classes = next(iter(dataloaders['val']))
print(inputs.shape)
# Make a grid from batch
out = torchvision.utils.make_grid(inputs)
imshow(out, title=[class_names[x] for x in classes])
```
### Create functions for training, validation, and evaluation
```
def train_model(model, criterion, optimizer, scheduler, num_epochs=25):
since = time.time()
best_model_wts = copy.deepcopy(model.state_dict())
best_acc = 0.0
num_img = {
"train": 0,
"val": 0,
}
datapoints_per_epoch = 100
imgs_per_datapoint = {
"train": int(float(dataset_sizes["train"] / datapoints_per_epoch)),
"val": int(float(dataset_sizes["val"] / datapoints_per_epoch)),
}
for epoch in range(num_epochs):
print('Epoch {}/{}'.format(epoch, num_epochs - 1))
print('-' * 10)
with open(f"/home/jupyter/logs/training/{type(model).__name__}-{since}.csv", "a") as f:
# Each epoch has a training and validation phase
for phase in ['train', 'val']:
if phase == 'train':
model.train() # Set model to training mode
else:
model.eval() # Set model to evaluate mode
running_loss = 0.0
running_corrects = 0
plot_loss = 0
plot_corrects = 0
# Iterate over data.
for inputs, labels in tqdm(dataloaders[phase], desc=f"Epoch: {epoch} ({phase})"):
inputs = inputs.to(device)
labels = labels.to(device)
# zero the parameter gradients
optimizer.zero_grad()
# forward
# track history if only in train
with torch.set_grad_enabled(phase == 'train'):
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# backward + optimize only if in training phase
if phase == 'train':
loss.backward()
optimizer.step()
# statistics
running_loss += loss.item() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
plot_loss += loss.item() * inputs.size(0)
plot_corrects += torch.sum(preds == labels.data)
num_img[phase] += BATCH_SIZE
if num_img[phase] % imgs_per_datapoint[phase] == 0:
f.write(f"{time.time()},{epoch},{phase},\
{num_img[phase]},{plot_loss / float(imgs_per_datapoint[phase])},\
{plot_corrects / float(imgs_per_datapoint[phase])}\n")
plot_loss = 0
plot_corrects = 0
"""
if phase == 'train':
scheduler.step()
"""
epoch_loss = running_loss / dataset_sizes[phase]
epoch_acc = running_corrects.double() / dataset_sizes[phase]
print('{} Loss: {:.4f} Acc: {:.4f}'.format(
phase, epoch_loss, epoch_acc))
# deep copy the model
if phase == 'val' and epoch_acc > best_acc:
best_acc = epoch_acc
best_model_wts = copy.deepcopy(model.state_dict())
torch.save(
{
"loss": epoch_loss,
"acc": epoch_acc,
"epoch": epoch,
"parameters": best_model_wts,
},
f"/home/jupyter/checkpoints/{type(model).__name__}-{since}.data",
)
print()
time_elapsed = time.time() - since
print('Training complete in {:.0f}m {:.0f}s'.format(
time_elapsed // 60, time_elapsed % 60))
print('Best val Acc: {:4f}'.format(best_acc))
# load best model weights
model.load_state_dict(best_model_wts)
return model
```
### Prepare and train the CNN
```
class CNN(nn.Module):
size_after_conv = 4 * 4 * 512
def __init__(self):
super(CNN, self).__init__()
self.features = nn.Sequential(
nn.Conv2d(1, 128, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
#nn.Dropout(),
nn.Conv2d(128, 256, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
#nn.Dropout(),
nn.Conv2d(256, 512, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Dropout(),
nn.Conv2d(512, 512, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Dropout(),
nn.Conv2d(512, 512, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Dropout(),
)
self.classify = nn.Sequential(
nn.Linear(self.size_after_conv, 2048),
nn.ReLU(),
nn.Dropout(),
nn.Linear(2048, 2048),
nn.ReLU(),
nn.Dropout(),
nn.Linear(2048, len(class_names)),
)
def forward(self, x):
x = self.features(x)
x = x.view(-1, self.size_after_conv)
x = self.classify(x)
return x
cnn = CNN().to(device)
print(cnn)
from collections import Counter
c = Counter()
for inputs, labels in dataloaders["train"]:
c.update(list(labels.numpy()))
c
class CNNSmall(nn.Module):
def __init__(self):
super(CNNSmall, self).__init__()
self.fc1 = nn.Linear(128*128, 1024)
self.fc2 = nn.Linear(1024, 1024)
self.fc3 = nn.Linear(1024, 4)
def forward(self, x):
x = x.view(-1, 128*128)
x = F.relu(self.fc1(x))
x = F.dropout(F.relu(self.fc2(x)), p=0.25)
x = F.relu(self.fc3(x))
return x
cnn2 = CNNSmall().to(device)
cnn2
class CNNSlightlyLarger(nn.Module):
size_after_conv = 4 * 4 * 32
def __init__(self):
super(CNNSlightlyLarger, self).__init__()
self.features = nn.Sequential(
nn.Conv2d(1, 32, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Conv2d(32, 32, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Conv2d(32, 32, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Conv2d(32, 32, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Conv2d(32, 32, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
)
self.classify = nn.Sequential(
nn.Linear(self.size_after_conv, 512),
nn.ReLU(),
nn.Linear(512, 512),
nn.ReLU(),
nn.Linear(512, len(class_names)),
)
def forward(self, x):
x = self.features(x)
x = x.view(-1, self.size_after_conv)
x = self.classify(x)
return x
cnn3 = CNNSlightlyLarger().to(device)
tensor_list = list(cnn2.state_dict().items())
for layer_tensor_name, tensor in tensor_list:
print('Layer {}: {} elements'.format(layer_tensor_name, torch.numel(tensor)))
print()
tensor_list = list(cnn.state_dict().items())
for layer_tensor_name, tensor in tensor_list:
print('Layer {}: {} elements'.format(layer_tensor_name, torch.numel(tensor)))
net = cnn2.cpu()
for idx, (inputs, labels) in enumerate(dataloaders["train"]):
print(labels)
# print(inputs)
out = F.softmax(net(inputs), dim=1)
print(out)
if idx > 1:
break
squeezenet = models.squeezenet1_1(pretrained=True)
num_chn = squeezenet.classifier[1].in_channels
squeezenet.classifier[1] = nn.Conv2d(num_chn, len(class_names), 1, 1)
squeezenet = squeezenet.to(device)
optimizer = torch.optim.Adam(squeezenet.classifier[1].parameters())
loss_function = nn.CrossEntropyLoss()
exp_lr_scheduler = lr_scheduler.StepLR(optimizer, step_size=7, gamma=0.1)
train_model(squeezenet, loss_function, optimizer, exp_lr_scheduler, num_epochs=25)
def visualize_model(model, num_images=6):
was_training = model.training
model.eval()
images_so_far = 0
fig = plt.figure()
with torch.no_grad():
for i, (inputs, labels) in enumerate(dataloaders['val']):
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
for j in range(inputs.size()[0]):
images_so_far += 1
ax = plt.subplot(num_images//2, 2, images_so_far)
ax.axis('off')
ax.set_title('predicted: {}'.format(class_names[preds[j]]))
imshow(inputs.cpu().data[j])
if images_so_far == num_images:
model.train(mode=was_training)
return
model.train(mode=was_training)
import matplotlib.pyplot as plt
images, labels = next(iter(test_loader))
for idx, (image, label) in enumerate(zip(images, labels)):
pred = int(torch.argmax(cnn(image.view(-1, 1, 64, 64).to(device))))
convert = {0: "Lars", 1: "Morgan", 2: "Kjartan", 3: "Ingen"}
plt.imshow(image.view(64, 64).cpu(), cmap="gray")
plt.text(2, 54, f"Image {idx + 1}", fontsize=14, color="white")
plt.text(2, 58, f"Predicted: `{convert[pred]}`", fontsize=14, color="white")
plt.text(2, 62, f"Actual : `{convert[label.item()]}`", fontsize=14, color="white")
plt.pause(0.05)
import cv2
cnn.eval()
class Label:
def __init__(self, label):
self.label = label
def item(self):
return self.label
filename = "/content/lars_5.png"
image = cv2.imread(filename, cv2.IMREAD_GRAYSCALE)
image = cv2.resize(image, (64, 64)) / 255.0
plt.imshow(image, cmap="gray")
convert = {0: "Lars", 1: "Morgan", 2: "Kjartan", 3: "Ingen"}
pred = int(torch.argmax(cnn(torch.Tensor(image).view(-1, 1, 64, 64).to(device))))
# plt.text(2, 58, f"Predicted: `{convert[pred]}`", fontsize=14, color="white")
# plt.text(2, 62, f"Actual : `{convert[label.item()]}`", fontsize=14, color="white")
if "morgan" in filename:
label = Label(1)
elif "lars" in filename:
label = Label(0)
else:
label = Label(3)
print(f"Predicted: `{convert[pred]}`" )
print(f"Actual : `{convert[label.item()]}`")
# plt.imshow(image.view(64, 64).cpu().to_numpy(), cmap="gray")
```
|
github_jupyter
|
gpu_info = !nvidia-smi
gpu_info = '\n'.join(gpu_info)
if gpu_info.find('failed') >= 0:
print('Select the Runtime > "Change runtime type" menu to enable a GPU accelerator, ')
print('and then re-execute this cell.')
else:
print(gpu_info)
from psutil import virtual_memory
ram_gb = virtual_memory().total / 1e9
print('Your runtime has {:.1f} gigabytes of available RAM\n'.format(ram_gb))
if ram_gb < 20:
print('To enable a high-RAM runtime, select the Runtime > "Change runtime type"')
print('menu, and then select High-RAM in the Runtime shape dropdown. Then, ')
print('re-execute this cell.')
else:
print('You are using a high-RAM runtime!')
import time
import os
import copy
import numpy as np
import matplotlib.pyplot as plt
from tqdm import tqdm
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.utils.data
from torch.optim import lr_scheduler
import torchvision
from torchvision import datasets, models, transforms
from google.cloud import storage
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
device
from google.cloud import storage
client = storage.Client()
bucket_name = "tdt4173-datasets"
bucket = client.get_bucket(bucket_name)
zipfilename = "/home/jupyter/data/faces/balanced_sampled_128px_color_130480_images_33_percent_val.zip"
blob_name = "faces/balanced_sampled_128px_color_130480_images_33_percent_val.zip"
blob = bucket.get_blob(blob_name)
blob.download_to_filename(zipfilename)
! rm -rf /home/jupyter/data/faces/images/val/.ipynb_checkpoints
!unzip /home/jupyter/data/faces/images_final_balanced_128px_color_26180_train_ca_300_val_per_class.zip -d /home/jupyter/data/faces/
BATCH_SIZE = 16
data_transforms = transforms.Compose([
transforms.Resize(224),
# transforms.CenterCrop(128),
transforms.Grayscale(num_output_channels=3),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225]),
# transforms.Normalize([0.5], [0.5]),
])
data_dir = '/home/jupyter/data/faces/images'
image_datasets = {x: datasets.ImageFolder(os.path.join(data_dir, x),
data_transforms)
for x in ['train', 'val']}
dataloaders = {x: torch.utils.data.DataLoader(image_datasets[x], batch_size=BATCH_SIZE,
shuffle=True, num_workers=4)
for x in ['train', 'val']}
dataset_sizes = {x: len(image_datasets[x]) for x in ['train', 'val']}
class_names = image_datasets['train'].classes
class_names
image_datasets['val'].classes
dataset_sizes
def imshow(inp, title=None):
"""Imshow for Tensor."""
inp = inp.numpy().transpose((1, 2, 0))
# mean = np.array([0.485, 0.456, 0.406])
# std = np.array([0.229, 0.224, 0.225])
mean = np.array([0.5])
std = np.array([0.5])
inp = std * inp + mean
inp = np.clip(inp, 0, 1)
plt.imshow(inp)
if title is not None:
plt.title(title)
plt.pause(0.001) # pause a bit so that plots are updated
# Get a batch of training data
inputs, classes = next(iter(dataloaders['val']))
print(inputs.shape)
# Make a grid from batch
out = torchvision.utils.make_grid(inputs)
imshow(out, title=[class_names[x] for x in classes])
def train_model(model, criterion, optimizer, scheduler, num_epochs=25):
since = time.time()
best_model_wts = copy.deepcopy(model.state_dict())
best_acc = 0.0
num_img = {
"train": 0,
"val": 0,
}
datapoints_per_epoch = 100
imgs_per_datapoint = {
"train": int(float(dataset_sizes["train"] / datapoints_per_epoch)),
"val": int(float(dataset_sizes["val"] / datapoints_per_epoch)),
}
for epoch in range(num_epochs):
print('Epoch {}/{}'.format(epoch, num_epochs - 1))
print('-' * 10)
with open(f"/home/jupyter/logs/training/{type(model).__name__}-{since}.csv", "a") as f:
# Each epoch has a training and validation phase
for phase in ['train', 'val']:
if phase == 'train':
model.train() # Set model to training mode
else:
model.eval() # Set model to evaluate mode
running_loss = 0.0
running_corrects = 0
plot_loss = 0
plot_corrects = 0
# Iterate over data.
for inputs, labels in tqdm(dataloaders[phase], desc=f"Epoch: {epoch} ({phase})"):
inputs = inputs.to(device)
labels = labels.to(device)
# zero the parameter gradients
optimizer.zero_grad()
# forward
# track history if only in train
with torch.set_grad_enabled(phase == 'train'):
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# backward + optimize only if in training phase
if phase == 'train':
loss.backward()
optimizer.step()
# statistics
running_loss += loss.item() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
plot_loss += loss.item() * inputs.size(0)
plot_corrects += torch.sum(preds == labels.data)
num_img[phase] += BATCH_SIZE
if num_img[phase] % imgs_per_datapoint[phase] == 0:
f.write(f"{time.time()},{epoch},{phase},\
{num_img[phase]},{plot_loss / float(imgs_per_datapoint[phase])},\
{plot_corrects / float(imgs_per_datapoint[phase])}\n")
plot_loss = 0
plot_corrects = 0
"""
if phase == 'train':
scheduler.step()
"""
epoch_loss = running_loss / dataset_sizes[phase]
epoch_acc = running_corrects.double() / dataset_sizes[phase]
print('{} Loss: {:.4f} Acc: {:.4f}'.format(
phase, epoch_loss, epoch_acc))
# deep copy the model
if phase == 'val' and epoch_acc > best_acc:
best_acc = epoch_acc
best_model_wts = copy.deepcopy(model.state_dict())
torch.save(
{
"loss": epoch_loss,
"acc": epoch_acc,
"epoch": epoch,
"parameters": best_model_wts,
},
f"/home/jupyter/checkpoints/{type(model).__name__}-{since}.data",
)
print()
time_elapsed = time.time() - since
print('Training complete in {:.0f}m {:.0f}s'.format(
time_elapsed // 60, time_elapsed % 60))
print('Best val Acc: {:4f}'.format(best_acc))
# load best model weights
model.load_state_dict(best_model_wts)
return model
class CNN(nn.Module):
size_after_conv = 4 * 4 * 512
def __init__(self):
super(CNN, self).__init__()
self.features = nn.Sequential(
nn.Conv2d(1, 128, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
#nn.Dropout(),
nn.Conv2d(128, 256, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
#nn.Dropout(),
nn.Conv2d(256, 512, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Dropout(),
nn.Conv2d(512, 512, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Dropout(),
nn.Conv2d(512, 512, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Dropout(),
)
self.classify = nn.Sequential(
nn.Linear(self.size_after_conv, 2048),
nn.ReLU(),
nn.Dropout(),
nn.Linear(2048, 2048),
nn.ReLU(),
nn.Dropout(),
nn.Linear(2048, len(class_names)),
)
def forward(self, x):
x = self.features(x)
x = x.view(-1, self.size_after_conv)
x = self.classify(x)
return x
cnn = CNN().to(device)
print(cnn)
from collections import Counter
c = Counter()
for inputs, labels in dataloaders["train"]:
c.update(list(labels.numpy()))
c
class CNNSmall(nn.Module):
def __init__(self):
super(CNNSmall, self).__init__()
self.fc1 = nn.Linear(128*128, 1024)
self.fc2 = nn.Linear(1024, 1024)
self.fc3 = nn.Linear(1024, 4)
def forward(self, x):
x = x.view(-1, 128*128)
x = F.relu(self.fc1(x))
x = F.dropout(F.relu(self.fc2(x)), p=0.25)
x = F.relu(self.fc3(x))
return x
cnn2 = CNNSmall().to(device)
cnn2
class CNNSlightlyLarger(nn.Module):
size_after_conv = 4 * 4 * 32
def __init__(self):
super(CNNSlightlyLarger, self).__init__()
self.features = nn.Sequential(
nn.Conv2d(1, 32, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Conv2d(32, 32, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Conv2d(32, 32, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Conv2d(32, 32, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
nn.Conv2d(32, 32, kernel_size=3, padding=1),
nn.MaxPool2d(2),
nn.ReLU(),
)
self.classify = nn.Sequential(
nn.Linear(self.size_after_conv, 512),
nn.ReLU(),
nn.Linear(512, 512),
nn.ReLU(),
nn.Linear(512, len(class_names)),
)
def forward(self, x):
x = self.features(x)
x = x.view(-1, self.size_after_conv)
x = self.classify(x)
return x
cnn3 = CNNSlightlyLarger().to(device)
tensor_list = list(cnn2.state_dict().items())
for layer_tensor_name, tensor in tensor_list:
print('Layer {}: {} elements'.format(layer_tensor_name, torch.numel(tensor)))
print()
tensor_list = list(cnn.state_dict().items())
for layer_tensor_name, tensor in tensor_list:
print('Layer {}: {} elements'.format(layer_tensor_name, torch.numel(tensor)))
net = cnn2.cpu()
for idx, (inputs, labels) in enumerate(dataloaders["train"]):
print(labels)
# print(inputs)
out = F.softmax(net(inputs), dim=1)
print(out)
if idx > 1:
break
squeezenet = models.squeezenet1_1(pretrained=True)
num_chn = squeezenet.classifier[1].in_channels
squeezenet.classifier[1] = nn.Conv2d(num_chn, len(class_names), 1, 1)
squeezenet = squeezenet.to(device)
optimizer = torch.optim.Adam(squeezenet.classifier[1].parameters())
loss_function = nn.CrossEntropyLoss()
exp_lr_scheduler = lr_scheduler.StepLR(optimizer, step_size=7, gamma=0.1)
train_model(squeezenet, loss_function, optimizer, exp_lr_scheduler, num_epochs=25)
def visualize_model(model, num_images=6):
was_training = model.training
model.eval()
images_so_far = 0
fig = plt.figure()
with torch.no_grad():
for i, (inputs, labels) in enumerate(dataloaders['val']):
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
for j in range(inputs.size()[0]):
images_so_far += 1
ax = plt.subplot(num_images//2, 2, images_so_far)
ax.axis('off')
ax.set_title('predicted: {}'.format(class_names[preds[j]]))
imshow(inputs.cpu().data[j])
if images_so_far == num_images:
model.train(mode=was_training)
return
model.train(mode=was_training)
import matplotlib.pyplot as plt
images, labels = next(iter(test_loader))
for idx, (image, label) in enumerate(zip(images, labels)):
pred = int(torch.argmax(cnn(image.view(-1, 1, 64, 64).to(device))))
convert = {0: "Lars", 1: "Morgan", 2: "Kjartan", 3: "Ingen"}
plt.imshow(image.view(64, 64).cpu(), cmap="gray")
plt.text(2, 54, f"Image {idx + 1}", fontsize=14, color="white")
plt.text(2, 58, f"Predicted: `{convert[pred]}`", fontsize=14, color="white")
plt.text(2, 62, f"Actual : `{convert[label.item()]}`", fontsize=14, color="white")
plt.pause(0.05)
import cv2
cnn.eval()
class Label:
def __init__(self, label):
self.label = label
def item(self):
return self.label
filename = "/content/lars_5.png"
image = cv2.imread(filename, cv2.IMREAD_GRAYSCALE)
image = cv2.resize(image, (64, 64)) / 255.0
plt.imshow(image, cmap="gray")
convert = {0: "Lars", 1: "Morgan", 2: "Kjartan", 3: "Ingen"}
pred = int(torch.argmax(cnn(torch.Tensor(image).view(-1, 1, 64, 64).to(device))))
# plt.text(2, 58, f"Predicted: `{convert[pred]}`", fontsize=14, color="white")
# plt.text(2, 62, f"Actual : `{convert[label.item()]}`", fontsize=14, color="white")
if "morgan" in filename:
label = Label(1)
elif "lars" in filename:
label = Label(0)
else:
label = Label(3)
print(f"Predicted: `{convert[pred]}`" )
print(f"Actual : `{convert[label.item()]}`")
# plt.imshow(image.view(64, 64).cpu().to_numpy(), cmap="gray")
| 0.698741 | 0.853791 |
# Control Flow
*Control flow* is where the rubber really meets the road in programming.
Without it, a program is simply a list of statements that are sequentially executed.
With control flow, you can execute certain code blocks conditionally and/or repeatedly: these basic building blocks can be combined to create surprisingly sophisticated programs!
Here we'll cover *conditional statements* (including "``if``", "``elif``", and "``else``"), *loop statements* (including "``for``", list comprehensions, and "``while``" and the accompanying "``break``" and "``continue``").
## Conditional Statements: ``if``-``elif``-``else``:
Conditional statements, often referred to as *if-then* statements, allow the programmer to execute certain pieces of code depending on some Boolean condition.
A basic example of a Python conditional statement is this:
```
x = -15
if x == 0:
print(x, "is zero")
elif x > 0:
print(x, "is positive")
elif x < 0:
print(x, "is negative")
else:
print(x, "is unlike anything I've ever seen...")
```
Note especially the use of colons (``:``) and whitespace to denote separate blocks of code.
Python adopts the ``if`` and ``else`` often used in other languages; its more unique keyword is ``elif``, a contraction of "else if".
In these conditional clauses, ``elif`` and ``else`` blocks are optional; additionally, you can optinally include as few or as many ``elif`` statements as you would like.
## ``for`` loops
Loops in Python are a way to repeatedly execute some code statement.
So, for example, if we'd like to print each of the items in a list, we can use a ``for`` loop:
```
for N in [2, 3, 5, 7]:
print(N, end=' ') # print all on same line
```
Notice the simplicity of the ``for`` loop: we specify the variable we want to use, the sequence we want to loop over, and use the "``in``" operator to link them together in an intuitive and readable way.
More precisely, the object to the right of the "``in``" can be any Python *iterator*.
An iterator can be thought of as a generalized sequence.
For example, one of the most commonly-used iterators in Python is the ``range`` object, which generates a sequence of numbers:
```
for i in range(10):
print(i, end=' ')
```
Note that the range starts at zero by default, and that by convention the top of the range is not included in the output.
Range objects can also have more complicated values:
```
# range from 5 to 10
list(range(5, 10))
# range from 0 to 10 by 2
list(range(0, 10, 2))
```
You might notice that the meaning of ``range`` arguments is very similar to the slicing syntax that we covered in Lists.
Note that the behavior of ``range()`` is one of the differences between Python 2 and Python 3: in Python 2, ``range()`` produces a list, while in Python 3, ``range()`` produces an iterable object.
## List Comprehensions
If you read enough Python code, you'll eventually come across the terse and efficient construction known as a *list comprehension*.
This is one feature of Python I expect you will fall in love with if you've not used it before; it looks something like this:
```
[i for i in range(20) if i % 3 > 0]
```
The result of this is a list of numbers which excludes multiples of 3.
While this example may seem a bit confusing at first, as familiarity with Python grows, reading and writing list comprehensions will become second nature.
### Basic List Comprehensions
List comprehensions are simply a way to compress a list-building for-loop into a single short, readable line.
For example, here is a loop that constructs a list of the first 12 square integers:
```
L = []
for n in range(12):
L.append(n ** 2)
L
```
The list comprehension equivalent of this is the following:
```
[n ** 2 for n in range(12)]
```
As with many Python statements, you can almost read-off the meaning of this statement in plain English: "construct a list consisting of the square of ``n`` for each ``n`` (from 0) up to 12".
This basic syntax, then, is ``[``*``expr(var)``* ``for`` *``var``* ``in`` *``iterable``*``]``, where *``expr(var)``* is any valid expression, *``var``* is a variable name, and *``iterable``* is any iterable Python object.
### Multiple Iteration
Sometimes you want to build a list not just from one value, but from two. To do this, simply add another ``for`` expression in the comprehension:
```
[(i, j) for i in range(2) for j in range(3)]
```
Notice that the second ``for`` expression acts as the interior index, varying the fastest in the resulting list.
This type of construction can be extended to three, four, or more iterators within the comprehension, though at some point code readibility will suffer!
### Conditionals on the Iterator
You can further control the iteration by adding a conditional to the end of the expression.
In the first example of the section, we iterated over all numbers from 1 to 20, but left-out multiples of 3.
Look at this again, and notice the construction:
```
[val for val in range(20) if val % 3 > 0]
```
The expression ``val % 3 > 0`` evaluates to ``True`` unless ``val`` is divisible by 3.
Again, the English language meaning can be immediately read off: "Construct a list of values for each value up to 20, but only if the value is not divisible by 3".
Once you are comfortable with it, this is much easier to write – and to understand at a glance – than the equivalent loop syntax:
```
L = []
for val in range(20):
if val % 3:
L.append(val)
L
```
### Conditionals on the Expression
If you've programmed in C, you might be familiar with the single-line conditional enabled by the ``?`` operator:
``` C
int absval = (val < 0) ? -val : val
```
Python has something very similar to this, which is most often used within list comprehensions, ``lambda`` functions, and other places where a simple expression is desired:
```
val = -10
val if val >= 0 else -val
```
We see that this simply duplicates the functionality of the built-in ``abs()`` function, but the construction lets you do some really interesting things within list comprehensions.
This is getting pretty complicated now, but you could do something like this:
```
[val if val % 2 else -val
for val in range(20) if val % 3]
```
Note the line break within the list comprehension before the ``for`` expression: this is valid in Python, and is often a nice way to break-up long list comprehensions for greater readibility.
Look this over: what we're doing is constructing a list, leaving out multiples of 3, and negating all mutliples of 2.
Once you understand the dynamics of list comprehensions, it's straightforward to move on to other types of comprehensions. The syntax is largely the same; the only difference is the type of bracket you use.
For example, with curly braces you can create a ``set`` with a *set comprehension*:
```
{n**2 for n in range(12)}
```
Recall that a ``set`` is a collection that contains no duplicates.
The set comprehension respects this rule, and eliminates any duplicate entries:
```
{a % 3 for a in range(1000)}
```
With a slight tweak, you can add a colon (``:``) to create a *dict comprehension*:
```
{("item-"+str(n)):n**2 for n in range(6)}
```
Finally, if you use parentheses rather than square brackets, you get what's called a *generator expression*:
```
(n**2 for n in range(12))
```
A generator expression is essentially a list comprehension in which elements are generated as-needed rather than all at-once, and the simplicity here belies the power of this language feature: we'll explore this more next.
## ``while`` loops
The other type of loop in Python is a ``while`` loop, which iterates until some condition is met:
```
i = 0
while i < 10:
print(i, end=' ')
i += 1
```
The argument of the ``while`` loop is evaluated as a boolean statement, and the loop is executed until the statement evaluates to False.
## ``break`` and ``continue``: Fine-Tuning Your Loops
There are two useful statements that can be used within loops to fine-tune how they are executed:
- The ``break`` statement breaks-out of the loop entirely
- The ``continue`` statement skips the remainder of the current loop, and goes to the next iteration
These can be used in both ``for`` and ``while`` loops.
Here is an example of using ``continue`` to print a string of odd numbers.
In this case, the result could be accomplished just as well with an ``if-else`` statement, but sometimes the ``continue`` statement can be a more convenient way to express the idea you have in mind:
```
for n in range(20):
# if the remainder of n / 2 is 0, skip the rest of the loop
if n % 2 == 0:
continue
print(n, end=' ')
```
Here is an example of a ``break`` statement used for a less trivial task.
This loop will fill a list with all Fibonacci numbers up to a certain value:
```
a, b = 0, 1
amax = 100
L = []
while True:
(a, b) = (b, a + b)
if a > amax:
break
L.append(a)
print(L)
```
Notice that we use a ``while True`` loop, which will loop forever unless we have a break statement!
## Loops with an ``else`` Block \*\*optional\*\*
One rarely used pattern available in Python is the ``else`` statement as part of a ``for`` or ``while`` loop.
We discussed the ``else`` block earlier: it executes if all the ``if`` and ``elif`` statements evaluate to ``False``.
The loop-``else`` is perhaps one of the more confusingly-named statements in Python; I prefer to think of it as a **no-break** statement: that is, the ``else`` block is executed only if the loop ends naturally, without encountering a ``break`` statement.
As an example of where this might be useful, consider the following (non-optimized) implementation of the *Sieve of Eratosthenes*, a well-known algorithm for finding prime numbers:
```
L = []
nmax = 30
for n in range(2, nmax):
for factor in L:
if n % factor == 0:
break
else: # no-break
L.append(n)
print(L)
```
The ``else`` statement only executes if none of the factors divide the given number.
The ``else`` statement works similarly with the ``while`` loop.
|
github_jupyter
|
x = -15
if x == 0:
print(x, "is zero")
elif x > 0:
print(x, "is positive")
elif x < 0:
print(x, "is negative")
else:
print(x, "is unlike anything I've ever seen...")
for N in [2, 3, 5, 7]:
print(N, end=' ') # print all on same line
for i in range(10):
print(i, end=' ')
# range from 5 to 10
list(range(5, 10))
# range from 0 to 10 by 2
list(range(0, 10, 2))
[i for i in range(20) if i % 3 > 0]
L = []
for n in range(12):
L.append(n ** 2)
L
[n ** 2 for n in range(12)]
[(i, j) for i in range(2) for j in range(3)]
[val for val in range(20) if val % 3 > 0]
L = []
for val in range(20):
if val % 3:
L.append(val)
L
Python has something very similar to this, which is most often used within list comprehensions, ``lambda`` functions, and other places where a simple expression is desired:
We see that this simply duplicates the functionality of the built-in ``abs()`` function, but the construction lets you do some really interesting things within list comprehensions.
This is getting pretty complicated now, but you could do something like this:
Note the line break within the list comprehension before the ``for`` expression: this is valid in Python, and is often a nice way to break-up long list comprehensions for greater readibility.
Look this over: what we're doing is constructing a list, leaving out multiples of 3, and negating all mutliples of 2.
Once you understand the dynamics of list comprehensions, it's straightforward to move on to other types of comprehensions. The syntax is largely the same; the only difference is the type of bracket you use.
For example, with curly braces you can create a ``set`` with a *set comprehension*:
Recall that a ``set`` is a collection that contains no duplicates.
The set comprehension respects this rule, and eliminates any duplicate entries:
With a slight tweak, you can add a colon (``:``) to create a *dict comprehension*:
Finally, if you use parentheses rather than square brackets, you get what's called a *generator expression*:
A generator expression is essentially a list comprehension in which elements are generated as-needed rather than all at-once, and the simplicity here belies the power of this language feature: we'll explore this more next.
## ``while`` loops
The other type of loop in Python is a ``while`` loop, which iterates until some condition is met:
The argument of the ``while`` loop is evaluated as a boolean statement, and the loop is executed until the statement evaluates to False.
## ``break`` and ``continue``: Fine-Tuning Your Loops
There are two useful statements that can be used within loops to fine-tune how they are executed:
- The ``break`` statement breaks-out of the loop entirely
- The ``continue`` statement skips the remainder of the current loop, and goes to the next iteration
These can be used in both ``for`` and ``while`` loops.
Here is an example of using ``continue`` to print a string of odd numbers.
In this case, the result could be accomplished just as well with an ``if-else`` statement, but sometimes the ``continue`` statement can be a more convenient way to express the idea you have in mind:
Here is an example of a ``break`` statement used for a less trivial task.
This loop will fill a list with all Fibonacci numbers up to a certain value:
Notice that we use a ``while True`` loop, which will loop forever unless we have a break statement!
## Loops with an ``else`` Block \*\*optional\*\*
One rarely used pattern available in Python is the ``else`` statement as part of a ``for`` or ``while`` loop.
We discussed the ``else`` block earlier: it executes if all the ``if`` and ``elif`` statements evaluate to ``False``.
The loop-``else`` is perhaps one of the more confusingly-named statements in Python; I prefer to think of it as a **no-break** statement: that is, the ``else`` block is executed only if the loop ends naturally, without encountering a ``break`` statement.
As an example of where this might be useful, consider the following (non-optimized) implementation of the *Sieve of Eratosthenes*, a well-known algorithm for finding prime numbers:
| 0.658637 | 0.971886 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
revenue_in_millions = [30990, 35119, 46542, 48017, 46854, 45998, 44294, 41863, 36212, 34300, 37266]
years = [2009, 2010, 2011, 2012, 2013, 2014, 2015, 2016, 2017, 2018,2019]
df = pd.DataFrame()
df['Year'] = years
df['Revenue'] = revenue_in_millions
df = df.set_index('Year')
df
from sklearn.preprocessing import StandardScaler
sc_x = StandardScaler()
x = df.index.values
x = x.reshape(len(x), 1)
x = sc_x.fit_transform(x)
sc_y = StandardScaler()
y = df['Revenue'].values
y = sc_y.fit_transform(y.reshape(len(y), 1))
from sklearn.svm import SVR
regressor = SVR(kernel = 'rbf',gamma='auto', tol=0.001, C=10.0, epsilon=0.001)
regressor.fit(x, y)
# plt.subplot(2,1,1)
# plt.plot(sc_x.inverse_transform(x), sc_y.inverse_transform(y)-sc_y.inverse_transform(y[0]), color = 'red', label = 'original')
#plt.plot(sc_x.inverse_transform(x), sc_y.inverse_transform(regressor.predict(x)), color = 'blue')
#plt.show()
#plt.subplot(2,1,2)
#plt.plot(sc_x.inverse_transform(x), sc_y.inverse_transform(regressor.predict(x)), color = 'blue')
xp = int(df.index.values[0])
xp = [x for x in range(xp,2023,1)]
xp = np.array(xp)
xp = np.reshape(xp,(-1,1))
#sc_xp = StandardScaler()
xp = sc_x.fit_transform(xp)
plt.plot(sc_x.inverse_transform(xp), sc_y.inverse_transform(regressor.predict(xp)), color = 'green', linestyle = '--', label = 'predicted')
plt.legend(loc='upper left')
plt.title('total revenue from sale')
plt.show()
print(sc_y.inverse_transform(y[0]),sc_y.inverse_transform(regressor.predict(sc_x.transform([[2022]]))))
#temp.append(sc_y.inverse_transform(regressor.predict(sc_x.transform([[2100]]))))
price_of_one = 5
plt.plot(sc_x.inverse_transform(xp), sc_y.inverse_transform(regressor.predict(xp))/price_of_one, color = 'green', linestyle = '--', label = 'predicted')
plt.legend(loc='upper left')
plt.title('number of cans sold')
plt.show()
dfs = pd.DataFrame()
dfs['Year'] = sc_x.inverse_transform(xp).ravel()
dfs['Revenue'] = sc_y.inverse_transform(regressor.predict(xp))
dfs = dfs.sort_values(by=['Revenue'])
dfs = dfs.reset_index(drop = True)
dfs
percentage_bottle_recycled = (1 - sc_y.inverse_transform(regressor.predict(xp))[-1]/dfs.iloc[-1,1])*100
percentage_bottle_recycled,dfs.iloc[-1,0]
amount_saved = percentage_bottle_recycled*max(sc_y.inverse_transform(regressor.predict(xp)))*0.26
amount_saved/1E+3
def amount_saved(x):
if x > 37746.754:
return (1 - sc_y.inverse_transform(regressor.predict(xp))[-1]/x)*x*26/1e3
else:
return None
dfs['Saved in kg'] = [amount_saved(x) for x in dfs['Revenue']]
dfs
dfs = dfs.sort_values(by=['Year'])
dfs = dfs.reset_index(drop=True)
dfs.to_csv('D:\Earth.Org\CokeRecycle\CocaCola_Plastic_Saved')
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
revenue_in_millions = [30990, 35119, 46542, 48017, 46854, 45998, 44294, 41863, 36212, 34300, 37266]
years = [2009, 2010, 2011, 2012, 2013, 2014, 2015, 2016, 2017, 2018,2019]
df = pd.DataFrame()
df['Year'] = years
df['Revenue'] = revenue_in_millions
df = df.set_index('Year')
df
from sklearn.preprocessing import StandardScaler
sc_x = StandardScaler()
x = df.index.values
x = x.reshape(len(x), 1)
x = sc_x.fit_transform(x)
sc_y = StandardScaler()
y = df['Revenue'].values
y = sc_y.fit_transform(y.reshape(len(y), 1))
from sklearn.svm import SVR
regressor = SVR(kernel = 'rbf',gamma='auto', tol=0.001, C=10.0, epsilon=0.001)
regressor.fit(x, y)
# plt.subplot(2,1,1)
# plt.plot(sc_x.inverse_transform(x), sc_y.inverse_transform(y)-sc_y.inverse_transform(y[0]), color = 'red', label = 'original')
#plt.plot(sc_x.inverse_transform(x), sc_y.inverse_transform(regressor.predict(x)), color = 'blue')
#plt.show()
#plt.subplot(2,1,2)
#plt.plot(sc_x.inverse_transform(x), sc_y.inverse_transform(regressor.predict(x)), color = 'blue')
xp = int(df.index.values[0])
xp = [x for x in range(xp,2023,1)]
xp = np.array(xp)
xp = np.reshape(xp,(-1,1))
#sc_xp = StandardScaler()
xp = sc_x.fit_transform(xp)
plt.plot(sc_x.inverse_transform(xp), sc_y.inverse_transform(regressor.predict(xp)), color = 'green', linestyle = '--', label = 'predicted')
plt.legend(loc='upper left')
plt.title('total revenue from sale')
plt.show()
print(sc_y.inverse_transform(y[0]),sc_y.inverse_transform(regressor.predict(sc_x.transform([[2022]]))))
#temp.append(sc_y.inverse_transform(regressor.predict(sc_x.transform([[2100]]))))
price_of_one = 5
plt.plot(sc_x.inverse_transform(xp), sc_y.inverse_transform(regressor.predict(xp))/price_of_one, color = 'green', linestyle = '--', label = 'predicted')
plt.legend(loc='upper left')
plt.title('number of cans sold')
plt.show()
dfs = pd.DataFrame()
dfs['Year'] = sc_x.inverse_transform(xp).ravel()
dfs['Revenue'] = sc_y.inverse_transform(regressor.predict(xp))
dfs = dfs.sort_values(by=['Revenue'])
dfs = dfs.reset_index(drop = True)
dfs
percentage_bottle_recycled = (1 - sc_y.inverse_transform(regressor.predict(xp))[-1]/dfs.iloc[-1,1])*100
percentage_bottle_recycled,dfs.iloc[-1,0]
amount_saved = percentage_bottle_recycled*max(sc_y.inverse_transform(regressor.predict(xp)))*0.26
amount_saved/1E+3
def amount_saved(x):
if x > 37746.754:
return (1 - sc_y.inverse_transform(regressor.predict(xp))[-1]/x)*x*26/1e3
else:
return None
dfs['Saved in kg'] = [amount_saved(x) for x in dfs['Revenue']]
dfs
dfs = dfs.sort_values(by=['Year'])
dfs = dfs.reset_index(drop=True)
dfs.to_csv('D:\Earth.Org\CokeRecycle\CocaCola_Plastic_Saved')
| 0.384103 | 0.634246 |
<img src="https://www.bestdesigns.co/uploads/inspiration_images/4350/990__1511457498_404_walmart.png" alt="WALMART LOGO" />
# Walmart : predict weekly sales
## Company's Description 📇
Walmart Inc. is an American multinational retail corporation that operates a chain of hypermarkets, discount department stores, and grocery stores from the United States, headquartered in Bentonville, Arkansas. The company was founded by Sam Walton in 1962.
## Project 🚧
Walmart's marketing service has asked you to build a machine learning model able to estimate the weekly sales in their stores, with the best precision possible on the predictions made. Such a model would help them understand better how the sales are influenced by economic indicators, and might be used to plan future marketing campaigns.
## Goals 🎯
The project can be divided into three steps:
- Part 1 : make an EDA and all the necessary preprocessings to prepare data for machine learning
- Part 2 : train a **linear regression model** (baseline)
- Part 3 : avoid overfitting by training a **regularized regression model**
## Scope of this project 🖼️
For this project, you'll work with a dataset that contains information about weekly sales achieved by different Walmart stores, and other variables such as the unemployment rate or the fuel price, that might be useful for predicting the amount of sales. The dataset has been taken from a Kaggle competition, but we made some changes compared to the original data. Please make sure that you're using **our** custom dataset (available on JULIE). 🤓
## Deliverable 📬
To complete this project, your team should:
- Create some visualizations
- Train at least one **linear regression model** on the dataset, that predicts the amount of weekly sales as a function of the other variables
- Assess the performances of the model by using a metric that is relevant for regression problems
- Interpret the coefficients of the model to identify what features are important for the prediction
- Train at least one model with **regularization (Lasso or Ridge)** to reduce overfitting
## Helpers 🦮
To help you achieve this project, here are a few tips that should help you:
### Part 1 : EDA and data preprocessing
Start your project by exploring your dataset : create figures, compute some statistics etc...
Then, you'll have to make some preprocessing on the dataset. You can follow the guidelines from the *preprocessing template*. There will also be some specific transformations to be planned on this dataset, for example on the *Date* column that can't be included as it is in the model. Below are some hints that might help you 🤓
#### Preprocessing to be planned with pandas
**Drop lines where target values are missing :**
- Here, the target variable (Y) corresponds to the column *Weekly_Sales*. One can see above that there are some missing values in this column.
- We never use imputation techniques on the target : it might create some bias in the predictions !
- Then, we will just drop the lines in the dataset for which the value in *Weekly_Sales* is missing.
**Create usable features from the *Date* column :**
The *Date* column cannot be included as it is in the model. Either you can drop this column, or you will create new columns that contain the following numeric features :
- *year*
- *month*
- *day*
- *day of week*
**Drop lines containing invalid values or outliers :**
In this project, will be considered as outliers all the numeric features that don't fall within the range : $[\bar{X} - 3\sigma, \bar{X} + 3\sigma]$. This concerns the columns : *Temperature*, *Fuel_price*, *CPI* and *Unemployment*
**Target variable/target (Y) that we will try to predict, to separate from the others** : *Weekly_Sales*
**------------**
#### Preprocessings to be planned with scikit-learn
**Explanatory variables (X)**
We need to identify which columns contain categorical variables and which columns contain numerical variables, as they will be treated differently.
- Categorical variables : Store, Holiday_Flag
- Numerical variables : Temperature, Fuel_Price, CPI, Unemployment, Year, Month, Day, DayOfWeek
### Part 2 : Baseline model (linear regression)
Once you've trained a first model, don't forget to assess its performances on the train and test sets. Are you satisfied with the results ?
Besides, it would be interesting to analyze the values of the model's coefficients to know what features are important for the prediction. To do so, the `.coef_` attribute of scikit-learn's LinearRegression class might be useful. Please refer to the following link for more information 😉 https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LinearRegression.html
### Part 3 : Fight overfitting
In this last part, you'll have to train a **regularized linear regression model**. You'll find below some useful classes in scikit-learn's documentation :
- https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Ridge.html#sklearn.linear_model.Ridge
- https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html#sklearn.linear_model.Lasso
**Bonus question**
In regularized regression models, there's a hyperparameter called *the regularization strength* that can be fine-tuned to get the best generalized predictions on a given dataset. This fine-tuning can be done thanks to scikit-learn's GridSearchCV class : https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html
Also, you'll find here some examples of how to use GridSearchCV together with Ridge or Lasso models : https://alfurka.github.io/2018-11-18-grid-search/
|
github_jupyter
|
<img src="https://www.bestdesigns.co/uploads/inspiration_images/4350/990__1511457498_404_walmart.png" alt="WALMART LOGO" />
# Walmart : predict weekly sales
## Company's Description 📇
Walmart Inc. is an American multinational retail corporation that operates a chain of hypermarkets, discount department stores, and grocery stores from the United States, headquartered in Bentonville, Arkansas. The company was founded by Sam Walton in 1962.
## Project 🚧
Walmart's marketing service has asked you to build a machine learning model able to estimate the weekly sales in their stores, with the best precision possible on the predictions made. Such a model would help them understand better how the sales are influenced by economic indicators, and might be used to plan future marketing campaigns.
## Goals 🎯
The project can be divided into three steps:
- Part 1 : make an EDA and all the necessary preprocessings to prepare data for machine learning
- Part 2 : train a **linear regression model** (baseline)
- Part 3 : avoid overfitting by training a **regularized regression model**
## Scope of this project 🖼️
For this project, you'll work with a dataset that contains information about weekly sales achieved by different Walmart stores, and other variables such as the unemployment rate or the fuel price, that might be useful for predicting the amount of sales. The dataset has been taken from a Kaggle competition, but we made some changes compared to the original data. Please make sure that you're using **our** custom dataset (available on JULIE). 🤓
## Deliverable 📬
To complete this project, your team should:
- Create some visualizations
- Train at least one **linear regression model** on the dataset, that predicts the amount of weekly sales as a function of the other variables
- Assess the performances of the model by using a metric that is relevant for regression problems
- Interpret the coefficients of the model to identify what features are important for the prediction
- Train at least one model with **regularization (Lasso or Ridge)** to reduce overfitting
## Helpers 🦮
To help you achieve this project, here are a few tips that should help you:
### Part 1 : EDA and data preprocessing
Start your project by exploring your dataset : create figures, compute some statistics etc...
Then, you'll have to make some preprocessing on the dataset. You can follow the guidelines from the *preprocessing template*. There will also be some specific transformations to be planned on this dataset, for example on the *Date* column that can't be included as it is in the model. Below are some hints that might help you 🤓
#### Preprocessing to be planned with pandas
**Drop lines where target values are missing :**
- Here, the target variable (Y) corresponds to the column *Weekly_Sales*. One can see above that there are some missing values in this column.
- We never use imputation techniques on the target : it might create some bias in the predictions !
- Then, we will just drop the lines in the dataset for which the value in *Weekly_Sales* is missing.
**Create usable features from the *Date* column :**
The *Date* column cannot be included as it is in the model. Either you can drop this column, or you will create new columns that contain the following numeric features :
- *year*
- *month*
- *day*
- *day of week*
**Drop lines containing invalid values or outliers :**
In this project, will be considered as outliers all the numeric features that don't fall within the range : $[\bar{X} - 3\sigma, \bar{X} + 3\sigma]$. This concerns the columns : *Temperature*, *Fuel_price*, *CPI* and *Unemployment*
**Target variable/target (Y) that we will try to predict, to separate from the others** : *Weekly_Sales*
**------------**
#### Preprocessings to be planned with scikit-learn
**Explanatory variables (X)**
We need to identify which columns contain categorical variables and which columns contain numerical variables, as they will be treated differently.
- Categorical variables : Store, Holiday_Flag
- Numerical variables : Temperature, Fuel_Price, CPI, Unemployment, Year, Month, Day, DayOfWeek
### Part 2 : Baseline model (linear regression)
Once you've trained a first model, don't forget to assess its performances on the train and test sets. Are you satisfied with the results ?
Besides, it would be interesting to analyze the values of the model's coefficients to know what features are important for the prediction. To do so, the `.coef_` attribute of scikit-learn's LinearRegression class might be useful. Please refer to the following link for more information 😉 https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LinearRegression.html
### Part 3 : Fight overfitting
In this last part, you'll have to train a **regularized linear regression model**. You'll find below some useful classes in scikit-learn's documentation :
- https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Ridge.html#sklearn.linear_model.Ridge
- https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html#sklearn.linear_model.Lasso
**Bonus question**
In regularized regression models, there's a hyperparameter called *the regularization strength* that can be fine-tuned to get the best generalized predictions on a given dataset. This fine-tuning can be done thanks to scikit-learn's GridSearchCV class : https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html
Also, you'll find here some examples of how to use GridSearchCV together with Ridge or Lasso models : https://alfurka.github.io/2018-11-18-grid-search/
| 0.810141 | 0.930899 |
# Simulated Sky Signal in time domain
In this lesson we will use the TOAST Operator `OpSimPySM` to create timestreams for an instrument given a sky model.
```
# Load common tools for all lessons
import sys
sys.path.insert(0, "..")
from lesson_tools import (
fake_focalplane
)
# Capture C++ output in the jupyter cells
%reload_ext wurlitzer
import toast
import healpy as hp
import numpy as np
env = toast.Environment.get()
env.set_log_level("DEBUG")
```
## Scanning strategy
Before being able to scan a map into a timestream we need to define a scanning strategy
and get pointing information for each channel.
We use the same **satellite** scanning used in lesson 2 about scanning strategies,
see the `02_Simulated_Scan_Strategies/simscan_satellite.ipynb` for more details.
```
focal_plane = fake_focalplane()
focal_plane.keys()
focal_plane["0A"]["fwhm_arcmin"]
# Scan parameters
alpha = 50.0 # precession opening angle, degrees
beta = 45.0 # spin opening angle, degrees
p_alpha = 25.0 # precession period, minutes
p_beta = 1.25 # spin period, minutes
samplerate = 0.5 # sample rate, Hz
hwprpm = 5.0 # HWP rotation in RPM
nside = 64 # Healpix NSIDE
# We will use one observation per day, with no gaps in between, and
# run for one year.
obs_samples = int(24 * 3600.0 * samplerate) - 1
nobs = 366
# Slew the precession axis so that it completes one circle
deg_per_day = 360.0 / nobs
from toast.todmap import TODSatellite, slew_precession_axis
detquat = {ch: focal_plane[ch]["quat"] for ch in focal_plane}
# Create distributed data
comm = toast.Comm()
data = toast.Data(comm)
# Append observations
for ob in range(nobs):
obsname = "{:03d}".format(ob)
obsfirst = ob * (obs_samples + 1)
obsstart = 24 * 3600.0
tod = TODSatellite(
comm.comm_group,
detquat,
obs_samples,
firstsamp=obsfirst,
firsttime=obsstart,
rate=samplerate,
spinperiod=p_beta,
spinangle=beta,
precperiod=p_alpha,
precangle=alpha,
coord="E",
hwprpm=hwprpm
)
qprec = np.empty(4 * tod.local_samples[1], dtype=np.float64).reshape((-1, 4))
slew_precession_axis(
qprec,
firstsamp=obsfirst,
samplerate=samplerate,
degday=deg_per_day,
)
tod.set_prec_axis(qprec=qprec)
obs = dict()
obs["tod"] = tod
data.obs.append(obs)
from toast.todmap import (
get_submaps_nested,
OpPointingHpix,
OpAccumDiag
)
from toast.map import (
DistPixels
)
# Make a simple pointing matrix
pointing = OpPointingHpix(nside=nside, nest=True, mode="IQU")
pointing.exec(data)
# Compute the locally hit pixels
localpix, localsm, subnpix = get_submaps_nested(data, nside)
# Construct a distributed map to store the hit map
npix = 12 * nside**2
hits = DistPixels(
comm=data.comm.comm_world,
size=npix,
nnz=1,
dtype=np.int64,
submap=subnpix,
local=localsm,
)
hits.data.fill(0)
# Accumulate the hit map locally
build_hits = OpAccumDiag(hits=hits)
build_hits.exec(data)
# Reduce the map across processes (a No-op in this case)
hits.allreduce()
%matplotlib inline
hp.mollview(hits.data.flatten(), nest=True)
```
## Define PySM parameters and instrument bandpasses
Then we define the sky model parameters, choosing the desired set of `PySM` models and then we specify the band center and the bandwidth for a top-hat bandpass.
Currently top-hat bandpasses are the only type supported by the operator, in the future we will implement arbitrary bandpasses.
Then bandpass parameters can be added directly to the `focal_plane` dictionary:
```
for ch in focal_plane:
focal_plane[ch]["bandcenter_ghz"] = 70
focal_plane[ch]["bandwidth_ghz"] = 10
focal_plane[ch]["fwhm"] = 60*2
pysm_sky_config = ["s1", "f1", "a1", "d1"] #syncrotron free free a&e and dust components of the sky
```
## Run the OpSimPySM operator
The `OpSimPySM` operator:
* Creates top-hat bandpasses arrays (frequency axis and weights) as expected by `PySM`
* Loops by channel and for each:
* Creates a `PySMSky` object just with 1 channel at a time
* Executes `PySMSky` to evaluate the sky models and bandpass-integrate
* Calls `PySM` to perform distributed smoothing with `libsharp`
* Gathers the map on the first MPI process
* Applies coordinate transformation if necessary (not currently implemented in `libsharp`)
* Use the `DistMap` object to communicate to each process the part of the sky they observe
* Calls `OpSimScan` to rescan the map to a timeline
```
from toast.todmap import OpSimPySM
OpSimPySM?
opsim_pysm = OpSimPySM(
comm=None,
pysm_model=pysm_sky_config,
nside=nside,
apply_beam=True,
debug=True,
focalplanes=[focal_plane],
subnpix=subnpix,
localsm=localsm
)
opsim_pysm.exec(data)
```
### Plot output timelines
```
%matplotlib inline
import matplotlib.pyplot as plt
tod = data.obs[0]['tod']
pix = tod.cache.reference("pixels_0A")
import toast.qarray as qa
theta, phi, pa = qa.to_angles(tod.read_pntg(detector="0A"))
#read_pntg gives quaternials and to angles gives theta the colatitude (0 = NP, 180 = SP) and phi and pa
pix
num = 10000
plt.figure(figsize=(7, 5))
plt.plot(np.degrees(theta[:num]), tod.cache.reference("signal_0A")[:num], ".")
plt.xlabel("$Colatitude [deg]$")
plt.ylabel("$Signal [ \mu K_{RJ} ]$");
num = 1000
plt.figure(figsize=(7, 5))
plt.plot(tod.cache.reference("signal_0A")[:num], "-")
plt.xlabel("$Time [arb.]$")
plt.ylabel("$Signal [ \mu K_{RJ} ]$");
#can see the signal as the pixel goes over the galaxy, another view of the same data
```
### Bin the output to a map
```
from numba import njit #just in time compiler for python can use this sometimes to avoid writting C++
@njit #causes numba to compile this function so that it runs faster
def just_make_me_a_map(output_map, signals):
"""Temperature only binner
Bins a list of (pix, signal) tuples into an output map,
it does not support polarization, so it just averages it out.
Parameters
----------
output_map : np.array
already zeroed output map
signals : numba.typed.List of (np.array[int64] pix, np.array[np.double] signal)
Returns
-------
hits : np.array[np.int64]
hitmap
"""
hits = np.zeros(len(output_map), dtype=np.int64)
for pix, signal in signals:
for p,s in zip(pix, signal):
output_map[p] += s
hits[p] += 1
output_map[hits != 0] /= hits[hits != 0]
return hits
from numba.typed import List
signals = List()
for obs in data.obs:
for ch in focal_plane:
signals.append((obs["tod"].cache.reference("pixels_%s" % ch), obs["tod"].cache.reference("signal_%s" % ch)))
output_map = np.zeros(npix, dtype=np.double)
h = just_make_me_a_map(output_map, signals)
hp.mollview(h, title="hitmap", nest=True)
hp.mollview(output_map, nest=True, min=0, max=1e-3, cmap="coolwarm") #making a map from our focal plane with 2 deg beams
hp.gnomview(output_map, rot=(0,0), xsize=5000, ysize=2000, cmap="coolwarm", nest=True, min=0, max=1e-2)
```
### Custom sky components
* `pysm_component_objects`: pass custom PySM component objects, see for example the [WebSkyCIB](https://so-pysm-models.readthedocs.io/en/latest/api/so_pysm_models.WebSkyCIB.html#so_pysm_models.WebSkyCIB) model in the [so_pysm_models](https://github.com/simonsobs/so_pysm_models) repository, it provides a Cosmic Infrared Background computed from
|
github_jupyter
|
# Load common tools for all lessons
import sys
sys.path.insert(0, "..")
from lesson_tools import (
fake_focalplane
)
# Capture C++ output in the jupyter cells
%reload_ext wurlitzer
import toast
import healpy as hp
import numpy as np
env = toast.Environment.get()
env.set_log_level("DEBUG")
focal_plane = fake_focalplane()
focal_plane.keys()
focal_plane["0A"]["fwhm_arcmin"]
# Scan parameters
alpha = 50.0 # precession opening angle, degrees
beta = 45.0 # spin opening angle, degrees
p_alpha = 25.0 # precession period, minutes
p_beta = 1.25 # spin period, minutes
samplerate = 0.5 # sample rate, Hz
hwprpm = 5.0 # HWP rotation in RPM
nside = 64 # Healpix NSIDE
# We will use one observation per day, with no gaps in between, and
# run for one year.
obs_samples = int(24 * 3600.0 * samplerate) - 1
nobs = 366
# Slew the precession axis so that it completes one circle
deg_per_day = 360.0 / nobs
from toast.todmap import TODSatellite, slew_precession_axis
detquat = {ch: focal_plane[ch]["quat"] for ch in focal_plane}
# Create distributed data
comm = toast.Comm()
data = toast.Data(comm)
# Append observations
for ob in range(nobs):
obsname = "{:03d}".format(ob)
obsfirst = ob * (obs_samples + 1)
obsstart = 24 * 3600.0
tod = TODSatellite(
comm.comm_group,
detquat,
obs_samples,
firstsamp=obsfirst,
firsttime=obsstart,
rate=samplerate,
spinperiod=p_beta,
spinangle=beta,
precperiod=p_alpha,
precangle=alpha,
coord="E",
hwprpm=hwprpm
)
qprec = np.empty(4 * tod.local_samples[1], dtype=np.float64).reshape((-1, 4))
slew_precession_axis(
qprec,
firstsamp=obsfirst,
samplerate=samplerate,
degday=deg_per_day,
)
tod.set_prec_axis(qprec=qprec)
obs = dict()
obs["tod"] = tod
data.obs.append(obs)
from toast.todmap import (
get_submaps_nested,
OpPointingHpix,
OpAccumDiag
)
from toast.map import (
DistPixels
)
# Make a simple pointing matrix
pointing = OpPointingHpix(nside=nside, nest=True, mode="IQU")
pointing.exec(data)
# Compute the locally hit pixels
localpix, localsm, subnpix = get_submaps_nested(data, nside)
# Construct a distributed map to store the hit map
npix = 12 * nside**2
hits = DistPixels(
comm=data.comm.comm_world,
size=npix,
nnz=1,
dtype=np.int64,
submap=subnpix,
local=localsm,
)
hits.data.fill(0)
# Accumulate the hit map locally
build_hits = OpAccumDiag(hits=hits)
build_hits.exec(data)
# Reduce the map across processes (a No-op in this case)
hits.allreduce()
%matplotlib inline
hp.mollview(hits.data.flatten(), nest=True)
for ch in focal_plane:
focal_plane[ch]["bandcenter_ghz"] = 70
focal_plane[ch]["bandwidth_ghz"] = 10
focal_plane[ch]["fwhm"] = 60*2
pysm_sky_config = ["s1", "f1", "a1", "d1"] #syncrotron free free a&e and dust components of the sky
from toast.todmap import OpSimPySM
OpSimPySM?
opsim_pysm = OpSimPySM(
comm=None,
pysm_model=pysm_sky_config,
nside=nside,
apply_beam=True,
debug=True,
focalplanes=[focal_plane],
subnpix=subnpix,
localsm=localsm
)
opsim_pysm.exec(data)
%matplotlib inline
import matplotlib.pyplot as plt
tod = data.obs[0]['tod']
pix = tod.cache.reference("pixels_0A")
import toast.qarray as qa
theta, phi, pa = qa.to_angles(tod.read_pntg(detector="0A"))
#read_pntg gives quaternials and to angles gives theta the colatitude (0 = NP, 180 = SP) and phi and pa
pix
num = 10000
plt.figure(figsize=(7, 5))
plt.plot(np.degrees(theta[:num]), tod.cache.reference("signal_0A")[:num], ".")
plt.xlabel("$Colatitude [deg]$")
plt.ylabel("$Signal [ \mu K_{RJ} ]$");
num = 1000
plt.figure(figsize=(7, 5))
plt.plot(tod.cache.reference("signal_0A")[:num], "-")
plt.xlabel("$Time [arb.]$")
plt.ylabel("$Signal [ \mu K_{RJ} ]$");
#can see the signal as the pixel goes over the galaxy, another view of the same data
from numba import njit #just in time compiler for python can use this sometimes to avoid writting C++
@njit #causes numba to compile this function so that it runs faster
def just_make_me_a_map(output_map, signals):
"""Temperature only binner
Bins a list of (pix, signal) tuples into an output map,
it does not support polarization, so it just averages it out.
Parameters
----------
output_map : np.array
already zeroed output map
signals : numba.typed.List of (np.array[int64] pix, np.array[np.double] signal)
Returns
-------
hits : np.array[np.int64]
hitmap
"""
hits = np.zeros(len(output_map), dtype=np.int64)
for pix, signal in signals:
for p,s in zip(pix, signal):
output_map[p] += s
hits[p] += 1
output_map[hits != 0] /= hits[hits != 0]
return hits
from numba.typed import List
signals = List()
for obs in data.obs:
for ch in focal_plane:
signals.append((obs["tod"].cache.reference("pixels_%s" % ch), obs["tod"].cache.reference("signal_%s" % ch)))
output_map = np.zeros(npix, dtype=np.double)
h = just_make_me_a_map(output_map, signals)
hp.mollview(h, title="hitmap", nest=True)
hp.mollview(output_map, nest=True, min=0, max=1e-3, cmap="coolwarm") #making a map from our focal plane with 2 deg beams
hp.gnomview(output_map, rot=(0,0), xsize=5000, ysize=2000, cmap="coolwarm", nest=True, min=0, max=1e-2)
| 0.65202 | 0.957158 |
# Lesson 1 Class Exercises: NumPy Part 1
```
import numpy as np
```
## Exercise #1
Write Python code to generate five random numbers from the normal distribution.
Look at the online documentation for [NumPy random number generators](https://docs.scipy.org/doc/numpy-1.15.0/reference/routines.random.html)
```
np.random.normal(loc=3, scale=2, size=5)
```
## Exercise #2
Write Python code to generate six random integers between 10 and 30
Look at the online documentation for [NumPy random number generators](https://docs.scipy.org/doc/numpy-1.15.0/reference/routines.random.html)
```
np.random.randint(low=10, high=30, size=6)
```
## Exercise #3
Write Python code to
+ create a 5x5 array with random values
+ print the array,
+ find and print the minimum and maximum values.
```
x = np.random.random((5,5))
print(x)
print(np.amin(x))
print(np.amax(x))
```
## Exercise #4
Write code using NumPy that would create a grid that could be used for the game of life. Follow these criteria
+ the grid must have 30 rows and 80 columns
+ it must be initalized to zeros.
+ print the grid dimensions to prove its size.
+ print the grid to show it is initalized to zeros.
```
y = np.zeros((30,80))
print(np.shape(y))
print(y)
```
## Exercise #5
Read the documentation about [genfromtxt()](https://docs.scipy.org/doc/numpy/reference/generated/numpy.genfromtxt.html) function of NumPy. What does it do? Do you understand what the arguments do?
A file has been provided to you named data2.txt. Be sure to place it in the same folder with this notebook. It contains the following text:
```
Value1,Value2,Value3
0.4839,0.4536,0.3561
0.1292,0.6875,NA
0.1781,0.3049,0.8928
NA,0.5801,0.2038
0.5993,0.4357,0.7410
```
Notice it has missing values. Write some code that
+ uses the `genfromtxt` function to read in this file into a numpy array.
+ prints the resulting array.
```
np.genfromtxt(fname='./data2.txt', dtype=float, delimiter=",", skip_header=1, missing_values=["NA"])
```
## Exercise #6
Write some code that
+ Creates an array of 100 rows and 10 columns filled with random values between 0 and 1
+ Print the first 10 rows.
+ Caculate the following statistics about each row and column
+ mean, variance, standard deviation and quartiles of each row.
+ print these values.
## Exercise #7
Write code that
+ Create an array of 100 rows and 10 columns of integer values with the minimum number 0 and the maximum 10,000
+ Print the first 10 rows of the matrix.
+ Calculate values for a histogram with 10 bins.
+ save in one variable the values of each bin
+ save in another variable the value at the left edge of the bin.
|
github_jupyter
|
import numpy as np
np.random.normal(loc=3, scale=2, size=5)
np.random.randint(low=10, high=30, size=6)
x = np.random.random((5,5))
print(x)
print(np.amin(x))
print(np.amax(x))
y = np.zeros((30,80))
print(np.shape(y))
print(y)
Value1,Value2,Value3
0.4839,0.4536,0.3561
0.1292,0.6875,NA
0.1781,0.3049,0.8928
NA,0.5801,0.2038
0.5993,0.4357,0.7410
np.genfromtxt(fname='./data2.txt', dtype=float, delimiter=",", skip_header=1, missing_values=["NA"])
| 0.157882 | 0.980839 |
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc" style="margin-top: 1em;"><ul class="toc-item"><li><span><a href="#Name" data-toc-modified-id="Name-1"><span class="toc-item-num">1 </span>Name</a></span></li><li><span><a href="#Search" data-toc-modified-id="Search-2"><span class="toc-item-num">2 </span>Search</a></span><ul class="toc-item"><li><span><a href="#Load-Cached-Results" data-toc-modified-id="Load-Cached-Results-2.1"><span class="toc-item-num">2.1 </span>Load Cached Results</a></span></li><li><span><a href="#Build-Model-From-Google-Images" data-toc-modified-id="Build-Model-From-Google-Images-2.2"><span class="toc-item-num">2.2 </span>Build Model From Google Images</a></span></li></ul></li><li><span><a href="#Analysis" data-toc-modified-id="Analysis-3"><span class="toc-item-num">3 </span>Analysis</a></span><ul class="toc-item"><li><span><a href="#Gender-cross-validation" data-toc-modified-id="Gender-cross-validation-3.1"><span class="toc-item-num">3.1 </span>Gender cross validation</a></span></li><li><span><a href="#Face-Sizes" data-toc-modified-id="Face-Sizes-3.2"><span class="toc-item-num">3.2 </span>Face Sizes</a></span></li><li><span><a href="#Screen-Time-Across-All-Shows" data-toc-modified-id="Screen-Time-Across-All-Shows-3.3"><span class="toc-item-num">3.3 </span>Screen Time Across All Shows</a></span></li><li><span><a href="#Appearances-on-a-Single-Show" data-toc-modified-id="Appearances-on-a-Single-Show-3.4"><span class="toc-item-num">3.4 </span>Appearances on a Single Show</a></span></li></ul></li><li><span><a href="#Persist-to-Cloud" data-toc-modified-id="Persist-to-Cloud-4"><span class="toc-item-num">4 </span>Persist to Cloud</a></span><ul class="toc-item"><li><span><a href="#Save-Model-to-Google-Cloud-Storage" data-toc-modified-id="Save-Model-to-Google-Cloud-Storage-4.1"><span class="toc-item-num">4.1 </span>Save Model to Google Cloud Storage</a></span></li><li><span><a href="#Save-Labels-to-DB" data-toc-modified-id="Save-Labels-to-DB-4.2"><span class="toc-item-num">4.2 </span>Save Labels to DB</a></span><ul class="toc-item"><li><span><a href="#Commit-the-person-and-labeler" data-toc-modified-id="Commit-the-person-and-labeler-4.2.1"><span class="toc-item-num">4.2.1 </span>Commit the person and labeler</a></span></li><li><span><a href="#Commit-the-FaceIdentity-labels" data-toc-modified-id="Commit-the-FaceIdentity-labels-4.2.2"><span class="toc-item-num">4.2.2 </span>Commit the FaceIdentity labels</a></span></li></ul></li></ul></li></ul></div>
```
from esper.prelude import *
from esper.identity import *
from esper import embed_google_images
```
# Name
Please add the person's name and their expected gender below (Male/Female).
```
name = 'Mika Brzezinski'
gender = 'Female'
```
# Search
## Load Cached Results
Reads cached identity model from local disk. Run this if the person has been labelled before and you only wish to regenerate the graphs. Otherwise, if you have never created a model for this person, please see the next section.
```
assert name != ''
results = FaceIdentityModel.load(name=name)
imshow(tile_imgs([cv2.resize(x[1][0], (200, 200)) for x in results.model_params['images']], cols=10))
plt.show()
plot_precision_and_cdf(results)
```
## Build Model From Google Images
Run this section if you do not have a cached model and precision curve estimates. This section will grab images using Google Image Search and score each of the faces in the dataset. We will interactively build the precision vs score curve.
It is important that the images that you select are accurate. If you make a mistake, rerun the cell below.
```
assert name != ''
# Grab face images from Google
img_dir = embed_google_images.fetch_images(name)
# If the images returned are not satisfactory, rerun the above with extra params:
# query_extras='' # additional keywords to add to search
# force=True # ignore cached images
face_imgs = load_and_select_faces_from_images(img_dir)
face_embs = embed_google_images.embed_images(face_imgs)
assert(len(face_embs) == len(face_imgs))
reference_imgs = tile_imgs([cv2.resize(x[0], (200, 200)) for x in face_imgs if x], cols=10)
def show_reference_imgs():
print('User selected reference images for {}.'.format(name))
imshow(reference_imgs)
plt.show()
show_reference_imgs()
# Score all of the faces in the dataset (this can take a minute)
face_ids_by_bucket, face_ids_to_score = face_search_by_embeddings(face_embs)
precision_model = PrecisionModel(face_ids_by_bucket)
```
Now we will validate which of the images in the dataset are of the target identity.
__Hover over with mouse and press S to select a face. Press F to expand the frame.__
```
show_reference_imgs()
print(('Mark all images that ARE NOT {}. Thumbnails are ordered by DESCENDING distance '
'to your selected images. (The first page is more likely to have non "{}" images.) '
'There are a total of {} frames. (CLICK THE DISABLE JUPYTER KEYBOARD BUTTON '
'BEFORE PROCEEDING.)').format(
name, name, precision_model.get_lower_count()))
lower_widget = precision_model.get_lower_widget()
lower_widget
show_reference_imgs()
print(('Mark all images that ARE {}. Thumbnails are ordered by ASCENDING distance '
'to your selected images. (The first page is more likely to have "{}" images.) '
'There are a total of {} frames. (CLICK THE DISABLE JUPYTER KEYBOARD BUTTON '
'BEFORE PROCEEDING.)').format(
name, name, precision_model.get_lower_count()))
upper_widget = precision_model.get_upper_widget()
upper_widget
```
Run the following cell after labelling to compute the precision curve. Do not forget to re-enable jupyter shortcuts.
```
# Compute the precision from the selections
lower_precision = precision_model.compute_precision_for_lower_buckets(lower_widget.selected)
upper_precision = precision_model.compute_precision_for_upper_buckets(upper_widget.selected)
precision_by_bucket = {**lower_precision, **upper_precision}
results = FaceIdentityModel(
name=name,
face_ids_by_bucket=face_ids_by_bucket,
face_ids_to_score=face_ids_to_score,
precision_by_bucket=precision_by_bucket,
model_params={
'images': list(zip(face_embs, face_imgs))
}
)
plot_precision_and_cdf(results)
```
The next cell persists the model locally.
```
results.save()
```
# Analysis
## Gender cross validation
Situations where the identity model disagrees with the gender classifier may be cause for alarm. We would like to check that instances of the person have the expected gender as a sanity check. This section shows the breakdown of the identity instances and their labels from the gender classifier.
```
gender_breakdown = compute_gender_breakdown(results)
print('Expected counts by gender:')
for k, v in gender_breakdown.items():
print(' {} : {}'.format(k, int(v)))
print()
print('Percentage by gender:')
denominator = sum(v for v in gender_breakdown.values())
for k, v in gender_breakdown.items():
print(' {} : {:0.1f}%'.format(k, 100 * v / denominator))
print()
```
Situations where the identity detector returns high confidence, but where the gender is not the expected gender indicate either an error on the part of the identity detector or the gender detector. The following visualization shows randomly sampled images, where the identity detector returns high confidence, grouped by the gender label.
```
high_probability_threshold = 0.8
show_gender_examples(results, high_probability_threshold)
```
## Face Sizes
Faces shown on-screen vary in size. For a person such as a host, they may be shown in a full body shot or as a face in a box. Faces in the background or those part of side graphics might be smaller than the rest. When calculuating screentime for a person, we would like to know whether the results represent the time the person was featured as opposed to merely in the background or as a tiny thumbnail in some graphic.
The next cell, plots the distribution of face sizes. Some possible anomalies include there only being very small faces or large faces.
```
plot_histogram_of_face_sizes(results)
```
The histogram above shows the distribution of face sizes, but not how those sizes occur in the dataset. For instance, one might ask why some faces are so large or whhether the small faces are actually errors. The following cell groups example faces, which are of the target identity with probability, by their sizes in terms of screen area.
```
high_probability_threshold = 0.8
show_faces_by_size(results, high_probability_threshold, n=10)
```
## Screen Time Across All Shows
One question that we might ask about a person is whether they received a significantly different amount of screentime on different shows. The following section visualizes the amount of screentime by show in total minutes and also in proportion of the show's total time. For a celebrity or political figure such as Donald Trump, we would expect significant screentime on many shows. For a show host such as Wolf Blitzer, we expect that the screentime be high for shows hosted by Wolf Blitzer.
```
screen_time_by_show = get_screen_time_by_show(results)
plot_screen_time_by_show(name, screen_time_by_show)
```
## Appearances on a Single Show
For people such as hosts, we would like to examine in greater detail the screen time allotted for a single show. First, fill in a show below.
```
show_name = 'Morning Joe'
# Compute the screen time for each video of the show
screen_time_by_video_id = compute_screen_time_by_video(results, show_name)
```
One question we might ask about a host is "how long they are show on screen" for an episode. Likewise, we might also ask for how many episodes is the host not present due to being on vacation or on assignment elsewhere. The following cell plots a histogram of the distribution of the length of the person's appearances in videos of the chosen show.
```
plot_histogram_of_screen_times_by_video(name, show_name, screen_time_by_video_id)
```
For a host, we expect screentime over time to be consistent as long as the person remains a host. For figures such as Hilary Clinton, we expect the screentime to track events in the real world such as the lead-up to 2016 election and then to drop afterwards. The following cell plots a time series of the person's screentime over time. Each dot is a video of the chosen show. Red Xs are videos for which the face detector did not run.
```
plot_screentime_over_time(name, show_name, screen_time_by_video_id)
```
We hypothesized that a host is more likely to appear at the beginning of a video and then also appear throughout the video. The following plot visualizes the distibution of shot beginning times for videos of the show.
```
plot_distribution_of_appearance_times_by_video(results, show_name)
```
# Persist to Cloud
The remaining code in this notebook uploads the built identity model to Google Cloud Storage and adds the FaceIdentity labels to the database.
## Save Model to Google Cloud Storage
```
gcs_model_path = results.save_to_gcs()
```
To ensure that the model stored to Google Cloud is valid, we load it and print the precision and cdf curve below.
```
gcs_results = FaceIdentityModel.load_from_gcs(name=name)
imshow(tile_imgs([cv2.resize(x[1][0], (200, 200)) for x in gcs_results.model_params['images']], cols=10))
plt.show()
plot_precision_and_cdf(gcs_results)
```
## Save Labels to DB
If you are satisfied with the model, we can commit the labels to the database.
```
from django.core.exceptions import ObjectDoesNotExist
def standardize_name(name):
return name.lower()
person_type = ThingType.objects.get(name='person')
try:
person = Thing.objects.get(name=standardize_name(name), type=person_type)
print('Found person:', person.name)
except ObjectDoesNotExist:
person = Thing(name=standardize_name(name), type=person_type)
print('Creating person:', person.name)
labeler = Labeler(name='face-identity-{}'.format(person.name), data_path=gcs_model_path)
```
### Commit the person and labeler
The labeler and person have been created but not set saved to the database. If a person was created, please make sure that the name is correct before saving.
```
person.save()
labeler.save()
```
### Commit the FaceIdentity labels
Now, we are ready to add the labels to the database. We will create a FaceIdentity for each face whose probability exceeds the minimum threshold.
```
commit_face_identities_to_db(results, person, labeler, min_threshold=0.001)
print('Committed {} labels to the db'.format(FaceIdentity.objects.filter(labeler=labeler).count()))
```
|
github_jupyter
|
from esper.prelude import *
from esper.identity import *
from esper import embed_google_images
name = 'Mika Brzezinski'
gender = 'Female'
assert name != ''
results = FaceIdentityModel.load(name=name)
imshow(tile_imgs([cv2.resize(x[1][0], (200, 200)) for x in results.model_params['images']], cols=10))
plt.show()
plot_precision_and_cdf(results)
assert name != ''
# Grab face images from Google
img_dir = embed_google_images.fetch_images(name)
# If the images returned are not satisfactory, rerun the above with extra params:
# query_extras='' # additional keywords to add to search
# force=True # ignore cached images
face_imgs = load_and_select_faces_from_images(img_dir)
face_embs = embed_google_images.embed_images(face_imgs)
assert(len(face_embs) == len(face_imgs))
reference_imgs = tile_imgs([cv2.resize(x[0], (200, 200)) for x in face_imgs if x], cols=10)
def show_reference_imgs():
print('User selected reference images for {}.'.format(name))
imshow(reference_imgs)
plt.show()
show_reference_imgs()
# Score all of the faces in the dataset (this can take a minute)
face_ids_by_bucket, face_ids_to_score = face_search_by_embeddings(face_embs)
precision_model = PrecisionModel(face_ids_by_bucket)
show_reference_imgs()
print(('Mark all images that ARE NOT {}. Thumbnails are ordered by DESCENDING distance '
'to your selected images. (The first page is more likely to have non "{}" images.) '
'There are a total of {} frames. (CLICK THE DISABLE JUPYTER KEYBOARD BUTTON '
'BEFORE PROCEEDING.)').format(
name, name, precision_model.get_lower_count()))
lower_widget = precision_model.get_lower_widget()
lower_widget
show_reference_imgs()
print(('Mark all images that ARE {}. Thumbnails are ordered by ASCENDING distance '
'to your selected images. (The first page is more likely to have "{}" images.) '
'There are a total of {} frames. (CLICK THE DISABLE JUPYTER KEYBOARD BUTTON '
'BEFORE PROCEEDING.)').format(
name, name, precision_model.get_lower_count()))
upper_widget = precision_model.get_upper_widget()
upper_widget
# Compute the precision from the selections
lower_precision = precision_model.compute_precision_for_lower_buckets(lower_widget.selected)
upper_precision = precision_model.compute_precision_for_upper_buckets(upper_widget.selected)
precision_by_bucket = {**lower_precision, **upper_precision}
results = FaceIdentityModel(
name=name,
face_ids_by_bucket=face_ids_by_bucket,
face_ids_to_score=face_ids_to_score,
precision_by_bucket=precision_by_bucket,
model_params={
'images': list(zip(face_embs, face_imgs))
}
)
plot_precision_and_cdf(results)
results.save()
gender_breakdown = compute_gender_breakdown(results)
print('Expected counts by gender:')
for k, v in gender_breakdown.items():
print(' {} : {}'.format(k, int(v)))
print()
print('Percentage by gender:')
denominator = sum(v for v in gender_breakdown.values())
for k, v in gender_breakdown.items():
print(' {} : {:0.1f}%'.format(k, 100 * v / denominator))
print()
high_probability_threshold = 0.8
show_gender_examples(results, high_probability_threshold)
plot_histogram_of_face_sizes(results)
high_probability_threshold = 0.8
show_faces_by_size(results, high_probability_threshold, n=10)
screen_time_by_show = get_screen_time_by_show(results)
plot_screen_time_by_show(name, screen_time_by_show)
show_name = 'Morning Joe'
# Compute the screen time for each video of the show
screen_time_by_video_id = compute_screen_time_by_video(results, show_name)
plot_histogram_of_screen_times_by_video(name, show_name, screen_time_by_video_id)
plot_screentime_over_time(name, show_name, screen_time_by_video_id)
plot_distribution_of_appearance_times_by_video(results, show_name)
gcs_model_path = results.save_to_gcs()
gcs_results = FaceIdentityModel.load_from_gcs(name=name)
imshow(tile_imgs([cv2.resize(x[1][0], (200, 200)) for x in gcs_results.model_params['images']], cols=10))
plt.show()
plot_precision_and_cdf(gcs_results)
from django.core.exceptions import ObjectDoesNotExist
def standardize_name(name):
return name.lower()
person_type = ThingType.objects.get(name='person')
try:
person = Thing.objects.get(name=standardize_name(name), type=person_type)
print('Found person:', person.name)
except ObjectDoesNotExist:
person = Thing(name=standardize_name(name), type=person_type)
print('Creating person:', person.name)
labeler = Labeler(name='face-identity-{}'.format(person.name), data_path=gcs_model_path)
person.save()
labeler.save()
commit_face_identities_to_db(results, person, labeler, min_threshold=0.001)
print('Committed {} labels to the db'.format(FaceIdentity.objects.filter(labeler=labeler).count()))
| 0.630116 | 0.964321 |
# Jupyter environment
The Jupyter is an open-source web application that allows you to create and share documents that contain live code, equations, visualizations and explanatory text. Uses include: data cleaning and transformation, numerical simulation, statistical modeling, machine learning and much more.
[Website](http://jupyter.org/) [Project repository](https://github.com/jupyter)
We will be using a Jupyter server as the primary web interface for this workshop. Several notebooks have been provided to you, in advance, to guide you through the workshop. After the workshop, you may use the [Agave Jupyter image](https://hub.docker.com/r/agaveplatform/jupyter-notebook/) to recreate the notebook server and repeat the workshop, or continue on with your own work at your leisure.
The Agave image has several customizations to facilitate use of the platform and ease much of the heavy lifting done behind the scenes in this tutorial.
### Custom Kernels
Your Jupyter server has multiple kernels available for use right away. We have preconfigured them with several useful libraries and tools to help users get up and running with common tasks easier. Additionally, we have bundled in Agave CLI and Python SDK into the Bash, Python 2, and Python 3 kernels respectively. Both kernels are pre-authenticated with valid Agave auth tokens that you can use to begin interacting with the Agave Platform right away.
### Shared file system
Your home directory on the Jupyter server is shared with your sandbox, so you can safely copy data between the two environments quickly and easily.
### Web console
Jupyter contains a web terminal that can be used to access your sandbox environment or interact with the Jupyter container itself. To login to your sandbox from the Jupyter web terminal, simply run the following command:
```
ssh -p 10022 $VM_IPADDRESS
```
### Tutorial notebooks
This tutorial is presented as a series of Jupyter notebooks. If you are attending this tutorial in person, you will download the notebooks into the home directory of your notebook server. If you are following along after the fact, you should download the notebooks from the github repository into your Jupyter workspace.
```
git clone --depth 1 https://github.com/agaveplatform/SC17-container-tutorial.git
```
### API access
The tutorial walks you through the process of obtaining a set of API keys an authenticating to the Agave Platform. Once this is done, you no longer need to authenticate to follow the tutorial. Both the Agave CLI and Python SDK will be picked up your authorization cache and automatically refresh it as needed.
### Extras
Inside of the `examples` directory, you will find several notebooks to help you learn more about the Agave platform, containers, and SciOps. We leave these for you to follow after the tutorial.
<hr>
# Sandbox environment
The tutorial sandbox is a full Ubuntu 16.04 server running as a Docker container on a VM dedicated for your use in this tutorial. The sandbox has a standard HPC build environment with OpenMPI, Python 2, Python 3, build-essential, gfortran, openssl, git, jq, vim, and a host of other utilities.
### Container runtimes
Docker and Singularity are both pre-installed in your Sandbox. All images used in this tutorial are available from the public Agave Docker Hub and Singularity Hub accounts. You may also use your own private registry accounts. You will need to login to the respective registries on your own.
### Funwave example code
The sample code for this project is already present in `$HOME/FUNWAVE-TVD`.
### Shared file system
Your `$HOME/work` directory on the Jupyter server is shared with your sandbox, so you can safely copy data between the two environments quickly and easily.
### Accessibility
To login to the sandbox from outside the Jupyter server, use the host IP address. You will find the public IP address of your sandbox in the `$VM_IPADDRESS` environment variable. Valid ssh keys are available in the `~/.ssh` director of your Jupyter server. Alternatively, you can append your own public key to the `$HOME/.ssh/authorized_keys` file.
```
ssh -i /path/to/private/key.pem -p 10022 jovyan@$VM_IPADDRESS
```
### Persistence
Your VM will remain available for 1-2 days following the tutorial. During that time, your data will remain available. After that, the VM an any data saved with it will be destroyed. If you need to persist your data, it is recommended that you move it to another host, or [create your own account](https://public.agaveapi.co/create_account) in the Agave public tenant and save your data in the free cloud storage provied to you by default there.
<hr>
# Logging In
We have already configured resources for you to use in this tutorial.
### Virtual Machine
Each of you have a dedicated VM provided by the [Nectar Cloud](https://nectar.org.au/cloudpage/). You will use this VM for the duration of the tutorial.
### Training Account
A training account on the Agave Platform's public tenant has also been allocated to you.
### Login
Your Jupyter server is available at `<username>.sc17.training.agaveplatform.org`.
Usernames will be training001 to training100. We will count off to determine our instance.
When you first login, you will find it empty, save for a notebook named [INSTALL.ipynb](INSTALL.ipynb)". Open this notebook by clicking on the notebook name, then click the *"run"* button. This will fetch all the tutorial notebooks from the tutorial's git repository an add them to your workspace.
Once complete, open the [Config](Config.ipynb) notebook to being the meat of our tutorial.
<hr>
# Following along at home
If you are following along with this tutorial at home, you can recreate the tutorial Jupyter server and sandbox environments by running the containers on your own server using the following Docker Compose file (i.e. save the file below in a file named `docker-compose.yml`).
```
version: '2'
volumes:
training-volume:
services:
jupyter:
image: agaveplatform/jupyter-notebook:latest
command: start-notebook.sh --NotebookApp.token=''
mem_limit: 2048m
ports:
- '8888:8005'
environment:
- VM_MACHINE=training-node-${AGAVE_USERNAME}
- VM_HOSTNAME=localhost:8888
- USE_TUNNEL=True
- ENVIRONMENT=training
- SCRATCH_DIR=/home/jovyan
- MACHINE_USERNAME=jovyan
- MACHINE_NAME=sandbox
- DOCKERHUB_NAME=stevenrbrandt
- AGAVE_APP_DEPLOYMENT_PATH=agave-deployment
- AGAVE_CACHE_DIR=/home/jovyan/work/.agave
- AGAVE_JSON_PARSER=jq
- AGAVE_USERNAME=${AGAVE_USERNAME}
- AGAVE_PASSWORD=${AGAVE_PASSWORD}
- AGAVE_SYSTEM_SITE_DOMAIN=localhost
- AGAVE_STORAGE_WORK_DIR=/home/jovyan
- AGAVE_STORAGE_HOME_DIR=/home/jovyan
- AGAVE_APP_NAME=funwave-tvd-sc17-${AGAVE_USERNAME}
- AGAVE_STORAGE_SYSTEM_ID=nectar-storage-${AGAVE_USERNAME}
- AGAVE_EXECUTION_SYSTEM_ID=nectar-exec${AGAVE_USERNAME}
volumes:
- training-volume:/home/jovyan/work
- ../notebooks:/home/jovyan/notebooks
sandbox:
image: agaveplatform/sc17-sandbox:latest
mem_limit: 2048m
privileged: True
ports:
- '10022:22'
environment:
- VM_MACHINE=training-node-${AGAVE_USERNAME}
- NGROK_TOKEN=${NGROK_TOKEN}
- USE_TUNNEL=True
- ENVIRONMENT=training
- AGAVE_CACHE_DIR=/home/jovyan/work/.agave
volumes:
- training-volume:/home/jovyan/work
- /var/run/docker.sock:/var/run/docker.sock
- $HOME/.docker:/home/jovyan/.docker:ro
```
> To run the above, you need to first set the environment variables `AGAVE_USERNAME`, `AGAVE_PASSWORD`, and `NGROK_TOKEN`. The first two should be your agave username and password as obtained from [Agave TOGO](https://togo.agaveapi.co). The ngrok token should be obtained from [ngrok](https://ngrok.com)
> Ngrok will provide tunnelling for you so that agave can ssh into your laptop or desktop machine. It will do this by setting the `M_IPADDRESS`, `VM_HOSTNAME` and `VM_SSH_PORT` for you.
> Once you have these things setup, you should be able to run `docker-compose up` (note: you should run this command from the same directory in which you created your `docker-compose.yml` file) you should then be able use your brower to connect to the tutorial setup on port 8888 of your local machine (http://localhost:8888).
|
github_jupyter
|
ssh -p 10022 $VM_IPADDRESS
git clone --depth 1 https://github.com/agaveplatform/SC17-container-tutorial.git
ssh -i /path/to/private/key.pem -p 10022 jovyan@$VM_IPADDRESS
version: '2'
volumes:
training-volume:
services:
jupyter:
image: agaveplatform/jupyter-notebook:latest
command: start-notebook.sh --NotebookApp.token=''
mem_limit: 2048m
ports:
- '8888:8005'
environment:
- VM_MACHINE=training-node-${AGAVE_USERNAME}
- VM_HOSTNAME=localhost:8888
- USE_TUNNEL=True
- ENVIRONMENT=training
- SCRATCH_DIR=/home/jovyan
- MACHINE_USERNAME=jovyan
- MACHINE_NAME=sandbox
- DOCKERHUB_NAME=stevenrbrandt
- AGAVE_APP_DEPLOYMENT_PATH=agave-deployment
- AGAVE_CACHE_DIR=/home/jovyan/work/.agave
- AGAVE_JSON_PARSER=jq
- AGAVE_USERNAME=${AGAVE_USERNAME}
- AGAVE_PASSWORD=${AGAVE_PASSWORD}
- AGAVE_SYSTEM_SITE_DOMAIN=localhost
- AGAVE_STORAGE_WORK_DIR=/home/jovyan
- AGAVE_STORAGE_HOME_DIR=/home/jovyan
- AGAVE_APP_NAME=funwave-tvd-sc17-${AGAVE_USERNAME}
- AGAVE_STORAGE_SYSTEM_ID=nectar-storage-${AGAVE_USERNAME}
- AGAVE_EXECUTION_SYSTEM_ID=nectar-exec${AGAVE_USERNAME}
volumes:
- training-volume:/home/jovyan/work
- ../notebooks:/home/jovyan/notebooks
sandbox:
image: agaveplatform/sc17-sandbox:latest
mem_limit: 2048m
privileged: True
ports:
- '10022:22'
environment:
- VM_MACHINE=training-node-${AGAVE_USERNAME}
- NGROK_TOKEN=${NGROK_TOKEN}
- USE_TUNNEL=True
- ENVIRONMENT=training
- AGAVE_CACHE_DIR=/home/jovyan/work/.agave
volumes:
- training-volume:/home/jovyan/work
- /var/run/docker.sock:/var/run/docker.sock
- $HOME/.docker:/home/jovyan/.docker:ro
| 0.227641 | 0.925129 |
```
# Imports
import numpy as np
import tensorflow as tf
from tensorflow.contrib import rnn
# Integer to Binary Generator
def getBin(integer, len_binary):
binary = bin(int(integer))[2:].zfill(len_binary)
return map(int, list(binary))
# Test case
number = 5.0
print getBin(number, 8)
# Dataset Creation
def create_data(num_samples, len_binary):
np.random.seed(1)
x = np.zeros(num_samples)
y = np.zeros((num_samples, len_binary))
max_val = 2 ** len_binary - 1
for i in range(num_samples):
number = np.random.randint(0, max_val)
x[i] = int(number)
y[i] = getBin(number, len_binary)
return x, y
# Test Case
X, y = create_data(5, 4)
for i in range(X.shape[0]):
print X[i], '\t ', y[i]
# TF Model Parameters
binary_length = 4
training_samples = 1000
testing_samples = 20
lr = 0.01
training_steps = 100000 # Need to train longer
display_steps = 5000
n_input = 1
n_hidden_units = 32 # Need more hidden units as compared to Binary to Int model
n_output = binary_length
timestep = 1
# Generate Training and Testing Data
X_train, y_train = create_data(training_samples, binary_length)
X_test, y_test = create_data(testing_samples, binary_length)
# Print data
display = 5
for i in range(display):
print X_train[i], '\t', y_train[i], "\n"
# TF Model and intializations
X = tf.placeholder(tf.float32, [None, timestep, n_input])
y = tf.placeholder(tf.float32, [None, n_output])
W = tf.Variable(tf.random_normal([n_hidden_units, n_output]))
b = tf.Variable(tf.random_normal([n_output]))
def model(X, W, b, timestep, n_hidden_units):
X = tf.unstack(X, timestep, 1)
lstm_cell = rnn.BasicLSTMCell(n_hidden_units, forget_bias=1.0)
outputs, states = rnn.static_rnn(lstm_cell, X, dtype=tf.float32)
logits = tf.matmul(outputs[-1], W) + b
return logits
logits = model(X, W, b, timestep, n_hidden_units)
loss = tf.reduce_mean(tf.losses.mean_squared_error(logits, y))
optimizer = tf.train.RMSPropOptimizer(lr)
training = optimizer.minimize(loss)
# Reshape data
X_train = np.reshape(X_train, [-1, timestep, n_input])
y_train = np.reshape(y_train, [-1, n_output])
X_test = np.reshape(X_test, [-1, timestep, n_input])
y_test = np.reshape(y_test, [-1, n_output])
# Print data
display = 5
for i in range(display):
print X_train[i], '\t', y_train[i]
# Run TF
np.random.seed(0)
with tf.Session() as sess:
tf.global_variables_initializer().run()
for step in range(training_steps):
_, loss_out = sess.run([training, loss], feed_dict={X: X_train, y:y_train})
if step % display_steps == 0:
print "Loss {} at timestep {}" .format(loss_out, step)
out = sess.run(logits, feed_dict={X: X_test})
# Evaluation Metric
# 1 if Prediction is greater than 0.5, 0 otherwise
mask = out > 0.5
out[mask] = 1
out[~mask]= 0
plot = True
if plot is True:
print "Ground Truth \t Predicted"
disp = 5
rdm = np.random.randint(0, y_test.shape[0], disp)
for i in rdm:
print y_test[i], "->", out[i]
acc = out == y_test
acc = acc.sum(axis=1) == binary_length
acc = acc.sum()/float(len(y_test))
print "Accuracy is {} \n" .format(acc)
```
|
github_jupyter
|
# Imports
import numpy as np
import tensorflow as tf
from tensorflow.contrib import rnn
# Integer to Binary Generator
def getBin(integer, len_binary):
binary = bin(int(integer))[2:].zfill(len_binary)
return map(int, list(binary))
# Test case
number = 5.0
print getBin(number, 8)
# Dataset Creation
def create_data(num_samples, len_binary):
np.random.seed(1)
x = np.zeros(num_samples)
y = np.zeros((num_samples, len_binary))
max_val = 2 ** len_binary - 1
for i in range(num_samples):
number = np.random.randint(0, max_val)
x[i] = int(number)
y[i] = getBin(number, len_binary)
return x, y
# Test Case
X, y = create_data(5, 4)
for i in range(X.shape[0]):
print X[i], '\t ', y[i]
# TF Model Parameters
binary_length = 4
training_samples = 1000
testing_samples = 20
lr = 0.01
training_steps = 100000 # Need to train longer
display_steps = 5000
n_input = 1
n_hidden_units = 32 # Need more hidden units as compared to Binary to Int model
n_output = binary_length
timestep = 1
# Generate Training and Testing Data
X_train, y_train = create_data(training_samples, binary_length)
X_test, y_test = create_data(testing_samples, binary_length)
# Print data
display = 5
for i in range(display):
print X_train[i], '\t', y_train[i], "\n"
# TF Model and intializations
X = tf.placeholder(tf.float32, [None, timestep, n_input])
y = tf.placeholder(tf.float32, [None, n_output])
W = tf.Variable(tf.random_normal([n_hidden_units, n_output]))
b = tf.Variable(tf.random_normal([n_output]))
def model(X, W, b, timestep, n_hidden_units):
X = tf.unstack(X, timestep, 1)
lstm_cell = rnn.BasicLSTMCell(n_hidden_units, forget_bias=1.0)
outputs, states = rnn.static_rnn(lstm_cell, X, dtype=tf.float32)
logits = tf.matmul(outputs[-1], W) + b
return logits
logits = model(X, W, b, timestep, n_hidden_units)
loss = tf.reduce_mean(tf.losses.mean_squared_error(logits, y))
optimizer = tf.train.RMSPropOptimizer(lr)
training = optimizer.minimize(loss)
# Reshape data
X_train = np.reshape(X_train, [-1, timestep, n_input])
y_train = np.reshape(y_train, [-1, n_output])
X_test = np.reshape(X_test, [-1, timestep, n_input])
y_test = np.reshape(y_test, [-1, n_output])
# Print data
display = 5
for i in range(display):
print X_train[i], '\t', y_train[i]
# Run TF
np.random.seed(0)
with tf.Session() as sess:
tf.global_variables_initializer().run()
for step in range(training_steps):
_, loss_out = sess.run([training, loss], feed_dict={X: X_train, y:y_train})
if step % display_steps == 0:
print "Loss {} at timestep {}" .format(loss_out, step)
out = sess.run(logits, feed_dict={X: X_test})
# Evaluation Metric
# 1 if Prediction is greater than 0.5, 0 otherwise
mask = out > 0.5
out[mask] = 1
out[~mask]= 0
plot = True
if plot is True:
print "Ground Truth \t Predicted"
disp = 5
rdm = np.random.randint(0, y_test.shape[0], disp)
for i in rdm:
print y_test[i], "->", out[i]
acc = out == y_test
acc = acc.sum(axis=1) == binary_length
acc = acc.sum()/float(len(y_test))
print "Accuracy is {} \n" .format(acc)
| 0.560373 | 0.63385 |
Setting up libcudnn7-doc (7.6.5.32-1+cuda10.0) ...This notebook will take a dataset of images, run them through TSNE to group them up (if enabled) then create a stylegan2 model with or without ADA.
Below are setting to choose when running this workflow. Make sure before running to have all images you want to use in a folder inside of the images folder. For example have a folder inside images called mona-lisa filled with pictures of different versions of the Mona Lisa. Please have the subfolder have no whitespaces in the name.
If TSNE is enable the program will halt after processing the images and ask you to choose which cluster to use. The clusters will be in the folder clusters.
Before running make sure your kernal is set to Python 3 (TensorFlow 1.15 Python 3.7 GPU Optimized)
```
dataset_name = 'mona-lisa'
use_ada = True
use_tsne = False
use_spacewalk = True
gpus = 2
# Crop Settings
# Choose center or no-crop
# TODO: Add random
crop_type = 'no-crop'
resolution = 512
# TSNE Settings
# Choose number of clusters to make or None for auto clustering
num_clusters = None
# ADA Settings
knum = 10
# Spacewalk Settings
fps = 24
seconds = 10
#Leave seeds = None for random seeds or
# enter a list in the form of [int, int, int..] to define the seeds
seeds = None
# set walk_type to 'line', 'sphere', 'noiseloop', or 'circularloop'
walk_type = 'sphere'
!pip install -r requirements.txt
import os
import train
from PIL import Image, ImageFile, ImageOps
import shutil
import math
ImageFile.LOAD_TRUNCATED_IMAGES = True
def resize(pil_img, res):
return pil_img.resize((res, res))
def crop_center(pil_img, res):
crop = res
img_width, img_height = pil_img.size
if img_width < crop:
crop = img_width
if img_height < crop:
crop = img_height
a = (img_width - crop) // 2
b = (img_height - crop) // 2
c = (img_width + crop) // 2
d = (img_height + crop) // 2
cropped_image = pil_img.crop((a,b,c,d))
return resize(cropped_image, res)
def no_crop(pil_img, res):
color = [0, 0, 0]
img_width, img_height = pil_img.size
if img_width < img_height:
top = 0
bottom = 0
left = math.ceil((img_height - img_width) / 2.0)
right = math.floor((img_height - img_width) / 2.0)
else:
top = math.ceil((img_height - img_width) / 2.0)
bottom = math.floor((img_height - img_width) / 2.0)
left = 0
right = 0
border_image = ImageOps.expand(pil_img, border=(left, top, right, bottom), fill='white')
return resize(border_image, res)
image_dir = './images/'
tmp_dir = './tmp/'
image_dir = os.path.join(image_dir, dataset_name)
tmp_dir = os.path.join(tmp_dir, dataset_name)
if not os.path.exists(tmp_dir):
os.makedirs(tmp_dir)
else:
try:
shutil.rmtree(tmp_dir)
except OSError as e:
print("Error: %s : %s" % (dir_path, e.strerror))
os.makedirs(tmp_dir)
for filename in os.listdir(image_dir):
file_extension = os.path.splitext(filename)[-1]
if file_extension != '.jpg' and file_extension != '.png':
print(file_extension)
continue
image_path = os.path.join(image_dir, filename)
image = Image.open(image_path)
mode = image.mode
if str(mode) != 'RGB':
continue
if crop_type == "center":
image = crop_center(image, resolution)
if crop_type == "no-crop":
image = no_crop(image, resolution)
tmp_path = os.path.join(tmp_dir, filename)
image.save(tmp_path)
if use_tsne:
!python tsne.py --path={tmp_dir}
else:
print('TSNE is not in use')
```
If TSNE is enabled when it is finished running check the Clusters folder and choose the cluster you want to use below
```
if use_tsne:
clusters = []
while True:
x = input("Enter a cluster you want to use or Enter to continue: ")
if x == '':
break
clusters.append(int(x))
dataset_dir = os.path.join("./datasets", dataset_name)
if use_ada and use_tsne:
image_dir = os.path.join("./tmp", str(dataset_name + "_clusters"))
!python dataset_tool.py create_from_images {dataset_dir} {image_dir}
!python train.py --outdir=./training-runs --gpus={gpus} --res={resolution} --data={dataset_dir} --kimg={knum}
elif use_ada:
image_dir = os.path.join("./tmp", dataset_name)
!python dataset_tool.py create_from_images {dataset_dir} {image_dir}
!python train.py --outdir=./training-runs --gpus={gpus} --res={resolution} --data={dataset_dir} --kimg={knum}
else:
print("ADA is not in use")
```
|
github_jupyter
|
dataset_name = 'mona-lisa'
use_ada = True
use_tsne = False
use_spacewalk = True
gpus = 2
# Crop Settings
# Choose center or no-crop
# TODO: Add random
crop_type = 'no-crop'
resolution = 512
# TSNE Settings
# Choose number of clusters to make or None for auto clustering
num_clusters = None
# ADA Settings
knum = 10
# Spacewalk Settings
fps = 24
seconds = 10
#Leave seeds = None for random seeds or
# enter a list in the form of [int, int, int..] to define the seeds
seeds = None
# set walk_type to 'line', 'sphere', 'noiseloop', or 'circularloop'
walk_type = 'sphere'
!pip install -r requirements.txt
import os
import train
from PIL import Image, ImageFile, ImageOps
import shutil
import math
ImageFile.LOAD_TRUNCATED_IMAGES = True
def resize(pil_img, res):
return pil_img.resize((res, res))
def crop_center(pil_img, res):
crop = res
img_width, img_height = pil_img.size
if img_width < crop:
crop = img_width
if img_height < crop:
crop = img_height
a = (img_width - crop) // 2
b = (img_height - crop) // 2
c = (img_width + crop) // 2
d = (img_height + crop) // 2
cropped_image = pil_img.crop((a,b,c,d))
return resize(cropped_image, res)
def no_crop(pil_img, res):
color = [0, 0, 0]
img_width, img_height = pil_img.size
if img_width < img_height:
top = 0
bottom = 0
left = math.ceil((img_height - img_width) / 2.0)
right = math.floor((img_height - img_width) / 2.0)
else:
top = math.ceil((img_height - img_width) / 2.0)
bottom = math.floor((img_height - img_width) / 2.0)
left = 0
right = 0
border_image = ImageOps.expand(pil_img, border=(left, top, right, bottom), fill='white')
return resize(border_image, res)
image_dir = './images/'
tmp_dir = './tmp/'
image_dir = os.path.join(image_dir, dataset_name)
tmp_dir = os.path.join(tmp_dir, dataset_name)
if not os.path.exists(tmp_dir):
os.makedirs(tmp_dir)
else:
try:
shutil.rmtree(tmp_dir)
except OSError as e:
print("Error: %s : %s" % (dir_path, e.strerror))
os.makedirs(tmp_dir)
for filename in os.listdir(image_dir):
file_extension = os.path.splitext(filename)[-1]
if file_extension != '.jpg' and file_extension != '.png':
print(file_extension)
continue
image_path = os.path.join(image_dir, filename)
image = Image.open(image_path)
mode = image.mode
if str(mode) != 'RGB':
continue
if crop_type == "center":
image = crop_center(image, resolution)
if crop_type == "no-crop":
image = no_crop(image, resolution)
tmp_path = os.path.join(tmp_dir, filename)
image.save(tmp_path)
if use_tsne:
!python tsne.py --path={tmp_dir}
else:
print('TSNE is not in use')
if use_tsne:
clusters = []
while True:
x = input("Enter a cluster you want to use or Enter to continue: ")
if x == '':
break
clusters.append(int(x))
dataset_dir = os.path.join("./datasets", dataset_name)
if use_ada and use_tsne:
image_dir = os.path.join("./tmp", str(dataset_name + "_clusters"))
!python dataset_tool.py create_from_images {dataset_dir} {image_dir}
!python train.py --outdir=./training-runs --gpus={gpus} --res={resolution} --data={dataset_dir} --kimg={knum}
elif use_ada:
image_dir = os.path.join("./tmp", dataset_name)
!python dataset_tool.py create_from_images {dataset_dir} {image_dir}
!python train.py --outdir=./training-runs --gpus={gpus} --res={resolution} --data={dataset_dir} --kimg={knum}
else:
print("ADA is not in use")
| 0.206094 | 0.726911 |
```
import numpy as np
from sklearn.multiclass import OneVsRestClassifier
from sklearn.metrics import precision_recall_fscore_support
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import accuracy_score
from sklearn.svm import SVC
import matplotlib.pyplot as plt
exp_dir = "./predict/0320_Dt_100_fine"
# exp_dir = "./predict/0320_Dt_80_fine"
# exp_dir = "./predict/0320_Dt_50_fine"
# exp_dir = "./predict/0320_Dt_30_fine"
# exp_dir = "./predict/0320_Dt_20_fine"
exp_dir = "./predict/0320_Dt_15_fine"
# exp_dir = "./predict/0320_Dt_10_fine"
# exp_dir = "./predict/0320_Dt_5_fine"
# exp_dir = "./predict/0320_Dt_1_fine"
# exp_dir = "./predict/0319_0"
# exp_dir = "./predict/0319_Dt_100_fine"
# exp_dir = "./predict/0319_Dst_100_fine"
# exp_dir = "./predict/0319_Dt_80_fine"
# exp_dir = "./predict/0319_Dst_80_fine"
# exp_dir = "./predict/0319_Dt_50_fine"
# exp_dir = "./predict/0319_Dst_50_fine"
# exp_dir = "./predict/0319_Dst_30_fine"
# exp_dir = "./predict/0319_Dt_30_fine"
# exp_dir = "./predict/0319_Dt_20_fine"
sX = np.loadtxt(f"{exp_dir}/Ds_train_100/vecs.tsv", dtype=np.float, delimiter='\t')
sY = np.loadtxt(f"{exp_dir}/Ds_train_100/metas.tsv", dtype=int, delimiter='\t')
sX = np.loadtxt(f"{exp_dir}/Ds_train/vecs.tsv", dtype=np.float, delimiter='\t')
sY = np.loadtxt(f"{exp_dir}/Ds_train/metas.tsv", dtype=int, delimiter='\t')
tX = np.loadtxt(f"{exp_dir}/Dt_train_100/vecs.tsv", dtype=np.float, delimiter='\t')
tY = np.loadtxt(f"{exp_dir}/Dt_train_100/metas.tsv", dtype=int, delimiter='\t')
tX = np.loadtxt(f"{exp_dir}/Dt_train/vecs.tsv", dtype=np.float, delimiter='\t')
tY = np.loadtxt(f"{exp_dir}/Dt_train/metas.tsv", dtype=int, delimiter='\t')
x_ = np.loadtxt(f"{exp_dir}/D_test/vecs.tsv", dtype=np.float, delimiter='\t')
y_ = np.loadtxt(f"{exp_dir}/D_test/metas.tsv", dtype=int, delimiter='\t')
x = np.loadtxt(f"{exp_dir}/D_test_new/vecs.tsv", dtype=np.float, delimiter='\t')
y = np.loadtxt(f"{exp_dir}/D_test_new/metas.tsv", dtype=int, delimiter='\t')
acc = list()
```
## Ds Domain: Precision, Recall, F1-score
### Test 100
```
X = sX
Y = sY
xt = x[:1400]
yt = y[:1400]
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(14)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(100) of the Ds domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
X = sX
Y = sY
xt = x_[:4201]
yt = y_[:4201]
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(14)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Ds domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
X = sX
Y = sY
xt = np.r_[x[:1400],x_[:4201]]
yt = np.r_[y[:1400],y_[:4201]]
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(14)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Ds domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
```
## Dt Domain: Precision, Recall, F1-score
```
X = tX
Y = tY
xt = x[1400:]
yt = y[1400:] - 14
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(5)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i+14) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(100) of the Dt domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
fig_, ax_ = plt.subplots(figsize=(10, 10))
label_font = {'size':'18'} # Adjust to fit
ax.set_title('Confusion Matrix of testing subset of Dt in ')
ax_.set_xlabel('Predicted labels', fontdict=label_font);
ax_.set_ylabel('Observed labels', fontdict=label_font);
plot_confusion_matrix(clf, xt, yt, values_format = 'd', ax=ax_)
X = tX
Y = tY
xt = x_[4201:]
yt = y_[4201:]-14
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(5)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i+14) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Dt domain')
plt.grid(True,axis='y')
ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
fig_, ax_ = plt.subplots(figsize=(10, 10))
label_font = {'size':'18'} # Adjust to fit
ax.set_title('Confusion Matrix of testing subset of Dt in ')
ax_.set_xlabel('Predicted labels', fontdict=label_font);
ax_.set_ylabel('Observed labels', fontdict=label_font);
plot_confusion_matrix(clf, xt, yt, values_format = 'd', ax=ax_)
X = tX
Y = tY
xt = np.r_[x[1401:],x_[4201:]]
yt = np.r_[y[1401:],y_[4201:]]-14
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(5)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Ds domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
fig_, ax_ = plt.subplots(figsize=(5, 5))
label_font = {'size':'12'} # Adjust to fit
ax_.set_title('Confusion Matrix of testing subset \nof Dt in 100-shot')
ax_.set_xlabel('Predicted labels', fontdict=label_font);
ax_.set_ylabel('Observed labels', fontdict=label_font);
plot_confusion_matrix(clf, xt, yt, values_format = 'd', ax=ax_)
```
## Ds + Dt Domain: Precision, Recall, F1-score
```
X = np.r_[sX,tX]
Y = np.r_[sY,tY+14]
xt = x
yt = y
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(19)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(100) of the Ds+Dt domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
X = np.r_[sX,tX]
Y = np.r_[sY,tY+14]
xt = x_
yt = y_
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(19)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Ds+Dt domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
X = np.r_[sX,tX]
Y = np.r_[sY,tY+14]
xt = np.r_[x,x_]
yt = np.r_[y,y_]
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(19)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Ds domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
fig_, ax_ = plt.subplots(figsize=(20, 20))
label_font = {'size':'18'} # Adjust to fit
ax_.set_xlabel('Predicted labels', fontdict=label_font);
ax_.set_ylabel('Observed labels', fontdict=label_font);
plot_confusion_matrix(clf, xt, yt, values_format = 'd', ax=ax_)
print(acc)
from matplotlib.ticker import FixedFormatter,FixedLocator
shot = [1,5,10,15,20,30,50,80,100]
ds_acc = [0.994822353,
0.984824139,
0.98428852,
0.985538297,
0.978575254,
0.978218175,
0.98036065,
0.980539189,
0.98428852]
dt_acc = [0.822322322,
0.857857858,
0.865865866,
0.916416416,
0.924424424,
0.92042042,
0.94044044,
0.953453453,
0.960960961]
dst_acc = [0.81,
0.890131579,
0.908289474,
0.924210526,
0.946052632,
0.944210526,
0.949868421,
0.954078947,
0.961447368]
fig, ax = plt.subplots()
ax.plot(shot, ds_acc, '--ro', label='Ds (14)')
ax.plot(shot, dt_acc, '--go', label='Dt (5)')
ax.plot(shot, dst_acc, '--bo', label='Ds+Dt (19)')
plt.grid(True,axis='y')
plt.xlabel("N-shot")
plt.ylabel("Accuracy (%)")
ax.xaxis.set_major_locator(FixedLocator(shot))
ax.xaxis.set_minor_locator(FixedLocator([i for i in range(20)]+[22,24,26,28,35,40,45,60,70,90]))
ax.legend(loc='lower right')
import matplotlib.pyplot as plt
from matplotlib.ticker import FixedFormatter,FixedLocator
shot = [0,1,5,10,15,20,30,50,80,100]
ds_acc = [0.993929655,
0.994108195,
0.985181218,
0.985002678,
0.985002678,
0.986966613,
0.985359757,
0.987502232,
0.985716836,
0.98410998]
dt_acc = [0.878878879,
0.928928929,
0.941441441,
0.937937938,
0.94044044,
0.941441441,
0.938438438,
0.951451451,
0.954954955,
0.95995996]
dst_acc = [0.918157895,
0.951184211,
0.950526316,
0.955394737,
0.959210526,
0.961315789,
0.960789474,
0.962631579,
0.961447368,
0.961447368,]
fig, ax = plt.subplots()
ax.plot(shot, ds_acc, '--ro', label='Ds (14)')
ax.plot(shot, dt_acc, '--go', label='Dt (5)')
ax.plot(shot, dst_acc, '--bo', label='Ds+Dt (19)')
plt.grid(True,axis='y')
plt.xlabel("N-shot")
plt.ylabel("Accuracy (%)")
ax.xaxis.set_major_locator(FixedLocator(shot))
ax.xaxis.set_minor_locator(FixedLocator([i for i in range(20)]+[22,24,26,28,35,40,45,60,70,90]))
ax.legend(loc='lower right')
from matplotlib.ticker import FixedFormatter,FixedLocator
shot = [0,1,5,10,15,20,30,50,80,100]
dt_acc = [0.672793097397778,
0.6702131023203266,
0.7792645449168972,
0.8143934247458903,
0.8414250676946725,
0.8396997203260183,
0.824126315101673,
0.8916387344189104,
0.9464370723172983,
0.9666899773945623]
fig, ax = plt.subplots()
ax.plot(shot, dt_acc, '--ro', label='Dt (14)')
plt.grid(True,axis='y')
plt.xlabel("N-shot")
plt.ylabel("Precision (%)")
ax.xaxis.set_major_locator(FixedLocator(shot))
ax.xaxis.set_minor_locator(FixedLocator([i for i in range(20)]+[22,24,26,28,35,40,45,60,70,90]))
# ax.legend(loc='lower right')
```
|
github_jupyter
|
import numpy as np
from sklearn.multiclass import OneVsRestClassifier
from sklearn.metrics import precision_recall_fscore_support
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import accuracy_score
from sklearn.svm import SVC
import matplotlib.pyplot as plt
exp_dir = "./predict/0320_Dt_100_fine"
# exp_dir = "./predict/0320_Dt_80_fine"
# exp_dir = "./predict/0320_Dt_50_fine"
# exp_dir = "./predict/0320_Dt_30_fine"
# exp_dir = "./predict/0320_Dt_20_fine"
exp_dir = "./predict/0320_Dt_15_fine"
# exp_dir = "./predict/0320_Dt_10_fine"
# exp_dir = "./predict/0320_Dt_5_fine"
# exp_dir = "./predict/0320_Dt_1_fine"
# exp_dir = "./predict/0319_0"
# exp_dir = "./predict/0319_Dt_100_fine"
# exp_dir = "./predict/0319_Dst_100_fine"
# exp_dir = "./predict/0319_Dt_80_fine"
# exp_dir = "./predict/0319_Dst_80_fine"
# exp_dir = "./predict/0319_Dt_50_fine"
# exp_dir = "./predict/0319_Dst_50_fine"
# exp_dir = "./predict/0319_Dst_30_fine"
# exp_dir = "./predict/0319_Dt_30_fine"
# exp_dir = "./predict/0319_Dt_20_fine"
sX = np.loadtxt(f"{exp_dir}/Ds_train_100/vecs.tsv", dtype=np.float, delimiter='\t')
sY = np.loadtxt(f"{exp_dir}/Ds_train_100/metas.tsv", dtype=int, delimiter='\t')
sX = np.loadtxt(f"{exp_dir}/Ds_train/vecs.tsv", dtype=np.float, delimiter='\t')
sY = np.loadtxt(f"{exp_dir}/Ds_train/metas.tsv", dtype=int, delimiter='\t')
tX = np.loadtxt(f"{exp_dir}/Dt_train_100/vecs.tsv", dtype=np.float, delimiter='\t')
tY = np.loadtxt(f"{exp_dir}/Dt_train_100/metas.tsv", dtype=int, delimiter='\t')
tX = np.loadtxt(f"{exp_dir}/Dt_train/vecs.tsv", dtype=np.float, delimiter='\t')
tY = np.loadtxt(f"{exp_dir}/Dt_train/metas.tsv", dtype=int, delimiter='\t')
x_ = np.loadtxt(f"{exp_dir}/D_test/vecs.tsv", dtype=np.float, delimiter='\t')
y_ = np.loadtxt(f"{exp_dir}/D_test/metas.tsv", dtype=int, delimiter='\t')
x = np.loadtxt(f"{exp_dir}/D_test_new/vecs.tsv", dtype=np.float, delimiter='\t')
y = np.loadtxt(f"{exp_dir}/D_test_new/metas.tsv", dtype=int, delimiter='\t')
acc = list()
X = sX
Y = sY
xt = x[:1400]
yt = y[:1400]
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(14)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(100) of the Ds domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
X = sX
Y = sY
xt = x_[:4201]
yt = y_[:4201]
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(14)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Ds domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
X = sX
Y = sY
xt = np.r_[x[:1400],x_[:4201]]
yt = np.r_[y[:1400],y_[:4201]]
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(14)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Ds domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
X = tX
Y = tY
xt = x[1400:]
yt = y[1400:] - 14
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(5)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i+14) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(100) of the Dt domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
fig_, ax_ = plt.subplots(figsize=(10, 10))
label_font = {'size':'18'} # Adjust to fit
ax.set_title('Confusion Matrix of testing subset of Dt in ')
ax_.set_xlabel('Predicted labels', fontdict=label_font);
ax_.set_ylabel('Observed labels', fontdict=label_font);
plot_confusion_matrix(clf, xt, yt, values_format = 'd', ax=ax_)
X = tX
Y = tY
xt = x_[4201:]
yt = y_[4201:]-14
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(5)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i+14) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Dt domain')
plt.grid(True,axis='y')
ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
fig_, ax_ = plt.subplots(figsize=(10, 10))
label_font = {'size':'18'} # Adjust to fit
ax.set_title('Confusion Matrix of testing subset of Dt in ')
ax_.set_xlabel('Predicted labels', fontdict=label_font);
ax_.set_ylabel('Observed labels', fontdict=label_font);
plot_confusion_matrix(clf, xt, yt, values_format = 'd', ax=ax_)
X = tX
Y = tY
xt = np.r_[x[1401:],x_[4201:]]
yt = np.r_[y[1401:],y_[4201:]]-14
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(5)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Ds domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
fig_, ax_ = plt.subplots(figsize=(5, 5))
label_font = {'size':'12'} # Adjust to fit
ax_.set_title('Confusion Matrix of testing subset \nof Dt in 100-shot')
ax_.set_xlabel('Predicted labels', fontdict=label_font);
ax_.set_ylabel('Observed labels', fontdict=label_font);
plot_confusion_matrix(clf, xt, yt, values_format = 'd', ax=ax_)
X = np.r_[sX,tX]
Y = np.r_[sY,tY+14]
xt = x
yt = y
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(19)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(100) of the Ds+Dt domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
X = np.r_[sX,tX]
Y = np.r_[sY,tY+14]
xt = x_
yt = y_
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(19)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Ds+Dt domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
X = np.r_[sX,tX]
Y = np.r_[sY,tY+14]
xt = np.r_[x,x_]
yt = np.r_[y,y_]
clf = SVC().fit(X, Y)
y_hat = clf.predict(xt)
result = precision_recall_fscore_support(yt, y_hat, average=None, labels=[i for i in range(19)])
print(accuracy_score(yt, y_hat))
acc.append(accuracy_score(yt, y_hat))
P = result[0]
R = result[1]
F = result[2]
label = [str(i) for i in F.argsort()]
P = P[F.argsort()]
R = R[F.argsort()]
F = F[F.argsort()]
fig, ax = plt.subplots() # Create a figure containing a single axes.
ax.plot(label, P, 'go', label='Precision')
ax.plot(label, R, 'ro', label='Recall')
ax.plot(label, F, '-bo', label='F1-score')
ax.set_title('Performance metric per class in test set(300) of the Ds domain')
plt.grid(True,axis='y')
# ax.set_ylim([0.5, 1.02])
ax.set_xticks(range(len(label)))
ax.set_xticklabels(label)
plt.xlabel("k(class number)")
plt.ylabel("Performance metric (%)")
ax.legend(loc='lower right')
fig_, ax_ = plt.subplots(figsize=(20, 20))
label_font = {'size':'18'} # Adjust to fit
ax_.set_xlabel('Predicted labels', fontdict=label_font);
ax_.set_ylabel('Observed labels', fontdict=label_font);
plot_confusion_matrix(clf, xt, yt, values_format = 'd', ax=ax_)
print(acc)
from matplotlib.ticker import FixedFormatter,FixedLocator
shot = [1,5,10,15,20,30,50,80,100]
ds_acc = [0.994822353,
0.984824139,
0.98428852,
0.985538297,
0.978575254,
0.978218175,
0.98036065,
0.980539189,
0.98428852]
dt_acc = [0.822322322,
0.857857858,
0.865865866,
0.916416416,
0.924424424,
0.92042042,
0.94044044,
0.953453453,
0.960960961]
dst_acc = [0.81,
0.890131579,
0.908289474,
0.924210526,
0.946052632,
0.944210526,
0.949868421,
0.954078947,
0.961447368]
fig, ax = plt.subplots()
ax.plot(shot, ds_acc, '--ro', label='Ds (14)')
ax.plot(shot, dt_acc, '--go', label='Dt (5)')
ax.plot(shot, dst_acc, '--bo', label='Ds+Dt (19)')
plt.grid(True,axis='y')
plt.xlabel("N-shot")
plt.ylabel("Accuracy (%)")
ax.xaxis.set_major_locator(FixedLocator(shot))
ax.xaxis.set_minor_locator(FixedLocator([i for i in range(20)]+[22,24,26,28,35,40,45,60,70,90]))
ax.legend(loc='lower right')
import matplotlib.pyplot as plt
from matplotlib.ticker import FixedFormatter,FixedLocator
shot = [0,1,5,10,15,20,30,50,80,100]
ds_acc = [0.993929655,
0.994108195,
0.985181218,
0.985002678,
0.985002678,
0.986966613,
0.985359757,
0.987502232,
0.985716836,
0.98410998]
dt_acc = [0.878878879,
0.928928929,
0.941441441,
0.937937938,
0.94044044,
0.941441441,
0.938438438,
0.951451451,
0.954954955,
0.95995996]
dst_acc = [0.918157895,
0.951184211,
0.950526316,
0.955394737,
0.959210526,
0.961315789,
0.960789474,
0.962631579,
0.961447368,
0.961447368,]
fig, ax = plt.subplots()
ax.plot(shot, ds_acc, '--ro', label='Ds (14)')
ax.plot(shot, dt_acc, '--go', label='Dt (5)')
ax.plot(shot, dst_acc, '--bo', label='Ds+Dt (19)')
plt.grid(True,axis='y')
plt.xlabel("N-shot")
plt.ylabel("Accuracy (%)")
ax.xaxis.set_major_locator(FixedLocator(shot))
ax.xaxis.set_minor_locator(FixedLocator([i for i in range(20)]+[22,24,26,28,35,40,45,60,70,90]))
ax.legend(loc='lower right')
from matplotlib.ticker import FixedFormatter,FixedLocator
shot = [0,1,5,10,15,20,30,50,80,100]
dt_acc = [0.672793097397778,
0.6702131023203266,
0.7792645449168972,
0.8143934247458903,
0.8414250676946725,
0.8396997203260183,
0.824126315101673,
0.8916387344189104,
0.9464370723172983,
0.9666899773945623]
fig, ax = plt.subplots()
ax.plot(shot, dt_acc, '--ro', label='Dt (14)')
plt.grid(True,axis='y')
plt.xlabel("N-shot")
plt.ylabel("Precision (%)")
ax.xaxis.set_major_locator(FixedLocator(shot))
ax.xaxis.set_minor_locator(FixedLocator([i for i in range(20)]+[22,24,26,28,35,40,45,60,70,90]))
# ax.legend(loc='lower right')
| 0.646683 | 0.703167 |
```
%matplotlib inline
import mir_eval, librosa, librosa.display, numpy, matplotlib.pyplot as plt, IPython.display as ipd
plt.style.use('seaborn-muted')
plt.rcParams['figure.figsize'] = (14, 5)
plt.rcParams['axes.grid'] = True
plt.rcParams['axes.spines.left'] = False
plt.rcParams['axes.spines.right'] = False
plt.rcParams['axes.spines.bottom'] = False
plt.rcParams['axes.spines.top'] = False
plt.rcParams['axes.xmargin'] = 0
plt.rcParams['axes.ymargin'] = 0
plt.rcParams['image.cmap'] = 'gray'
plt.rcParams['image.interpolation'] = None
```
[← Back to Index](index.html)
# Evaluation Example: Beat Tracking
[Documentation: `mir_eval.beat`](http://craffel.github.io/mir_eval/#module-mir_eval.beat)
Evaluation method: compute the error between the estimated beat times and some reference list of beat locations. Many metrics additionally compare the beat sequences at different metric levels in order to deal with the ambiguity of tempo.
Let's evaluate a beat detector on the following audio:
```
y, sr = librosa.load('audio/prelude_cmaj.wav')
ipd.Audio(y, rate=sr)
```
## Detect Beats
Estimate the beats using `beat_track`:
```
est_tempo, est_beats = librosa.beat.beat_track(y=y, sr=sr, bpm=120)
est_beats = librosa.frames_to_time(est_beats, sr=sr)
est_beats
```
Load a fictional reference annotation.
```
ref_beats = numpy.array([0, 0.50, 1.02, 1.53, 1.99, 2.48, 2.97,
3.43, 3.90, 4.41, 4.89, 5.38,
5.85, 6.33, 6.82, 7.29, 7.70])
```
Plot the estimated and reference beats together.
```
D = librosa.stft(y)
S = abs(D)
S_db = librosa.amplitude_to_db(S)
librosa.display.specshow(S_db, sr=sr, x_axis='time', y_axis='log')
plt.ylim(0, 8192)
plt.vlines(est_beats, 0, 8192, color='#00ff00')
plt.scatter(ref_beats, 5000*numpy.ones_like(ref_beats), color='k', s=100)
```
## Evaluate
Evaluate using [`mir_eval.beat.evaluate`](https://github.com/craffel/mir_eval/blob/master/mir_eval/beat.py#L704):
```
mir_eval.beat.evaluate(ref_beats, est_beats)
```
## Example: Chord Estimation
```
mir_eval.chord.evaluate()
```
Hidden benefits
- Input validation! Many errors can be traced back to ill-formatted data.
- Standardized behavior, full test coverage.
## More than metrics
mir_eval has tools for display and sonification.
```
import librosa.display
import mir_eval.display
```
Common plots: `events`, `labeled_intervals`
pitch, multipitch, piano_roll
segments, hierarchy,
separation
### Example: Events
```
librosa.display.specshow(S, x_axis='time', y_axis='mel')
mir_eval.display.events(ref_beats, color='w', alpha=0.8, linewidth=3)
mir_eval.display.events(est_beats, color='c', alpha=0.8, linewidth=3, linestyle='--')
```
### Example: Labeled Intervals
### Example: Source Separation
```
y_harm, y_perc = librosa.effects.hpss(y, margin=8)
plt.figure(figsize=(12, 4))
mir_eval.display.separation([y_perc, y_harm], sr, labels=['percussive', 'harmonic'])
plt.legend()
Audio(data=numpy.vstack([
mir_eval.sonify.chords()
```
[← Back to Index](index.html)
|
github_jupyter
|
%matplotlib inline
import mir_eval, librosa, librosa.display, numpy, matplotlib.pyplot as plt, IPython.display as ipd
plt.style.use('seaborn-muted')
plt.rcParams['figure.figsize'] = (14, 5)
plt.rcParams['axes.grid'] = True
plt.rcParams['axes.spines.left'] = False
plt.rcParams['axes.spines.right'] = False
plt.rcParams['axes.spines.bottom'] = False
plt.rcParams['axes.spines.top'] = False
plt.rcParams['axes.xmargin'] = 0
plt.rcParams['axes.ymargin'] = 0
plt.rcParams['image.cmap'] = 'gray'
plt.rcParams['image.interpolation'] = None
y, sr = librosa.load('audio/prelude_cmaj.wav')
ipd.Audio(y, rate=sr)
est_tempo, est_beats = librosa.beat.beat_track(y=y, sr=sr, bpm=120)
est_beats = librosa.frames_to_time(est_beats, sr=sr)
est_beats
ref_beats = numpy.array([0, 0.50, 1.02, 1.53, 1.99, 2.48, 2.97,
3.43, 3.90, 4.41, 4.89, 5.38,
5.85, 6.33, 6.82, 7.29, 7.70])
D = librosa.stft(y)
S = abs(D)
S_db = librosa.amplitude_to_db(S)
librosa.display.specshow(S_db, sr=sr, x_axis='time', y_axis='log')
plt.ylim(0, 8192)
plt.vlines(est_beats, 0, 8192, color='#00ff00')
plt.scatter(ref_beats, 5000*numpy.ones_like(ref_beats), color='k', s=100)
mir_eval.beat.evaluate(ref_beats, est_beats)
mir_eval.chord.evaluate()
import librosa.display
import mir_eval.display
librosa.display.specshow(S, x_axis='time', y_axis='mel')
mir_eval.display.events(ref_beats, color='w', alpha=0.8, linewidth=3)
mir_eval.display.events(est_beats, color='c', alpha=0.8, linewidth=3, linestyle='--')
y_harm, y_perc = librosa.effects.hpss(y, margin=8)
plt.figure(figsize=(12, 4))
mir_eval.display.separation([y_perc, y_harm], sr, labels=['percussive', 'harmonic'])
plt.legend()
Audio(data=numpy.vstack([
mir_eval.sonify.chords()
| 0.406273 | 0.907271 |
# Symmetrize
In this notebook, we will use the _symmetrize_ function to create bi-directional edges in an undirected graph
Notebook Credits
* Original Authors: Bradley Rees and James Wyles
* Created: 08/13/2019
* Updated: 03/02/2020
RAPIDS Versions: 0.13
Test Hardware
* GV100 32G, CUDA 10.2
## Introduction
In many cases, an Undirected graph is saved as a single edge between vertex pairs. That saves a lot of space in the data file. However, in order to process that data in cuGraph, there needs to be an edge in each direction for undirected. Converting from a single edge to two edges, one in each direction, is called symmetrization.
To symmetrize an edge list (COO data) use:<br>
**cugraph.symmetrize(source, destination, value)**
* __source__: cudf.Series
* __destination__: cudf.Series
* __value__: cudf.Series
Returns:
* __triplet__: three variables are returned:
* __source__: cudf.Series
* __destination__: cudf.Series
* __value__: cudf.Series
### Test Data
We will be using an undirected unsymmetrized version of the Zachary Karate club dataset. The result of symmetrization should be a dataset equal to to the version used in the PageRank notebook.
*W. W. Zachary, An information flow model for conflict and fission in small groups, Journal of
Anthropological Research 33, 452-473 (1977).*

```
# Import needed libraries
import cugraph
import cudf
# Read the unsymmetrized data
unsym_data ='../data/karate_undirected.csv'
gdf = cudf.read_csv(unsym_data, names=["src", "dst"], delimiter='\t', dtype=["int32", "int32"] )
# load the full symmetrized dataset for comparison
datafile='../data/karate-data.csv'
test_gdf = cudf.read_csv(datafile, names=["src", "dst"], delimiter='\t', dtype=["int32", "int32"] )
print("Unsymmetrized Graph")
print("\tNumber of Edges: " + str(len(gdf)))
print("Baseline Graph")
print("\tNumber of Edges: " + str(len(test_gdf)))
```
_Since the unsymmetrized graph only has one edge between vertices, that underlying code treats that as a directed graph_
```
G = cugraph.Graph()
G.from_cudf_edgelist(gdf, source='src', destination='dst')
gdf_page = cugraph.pagerank(G)
# best PR score is
m = gdf_page['pagerank'].max()
df = gdf_page.query('pagerank == @m')
df
```
### Now Symmetrize the dataset
```
sdf = cugraph.symmetrize_df(gdf, 'src', 'dst')
print("Unsymmetrized Graph")
print("\tNumber of Edges: " + str(len(gdf)))
print("Symmetrized Graph")
print("\tNumber of Edges: " + str(len(sdf)))
print("Baseline Graph")
print("\tNumber of Edges: " + str(len(test_gdf)))
```
---
Copyright (c) 2019-2020, NVIDIA CORPORATION.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
___
|
github_jupyter
|
# Import needed libraries
import cugraph
import cudf
# Read the unsymmetrized data
unsym_data ='../data/karate_undirected.csv'
gdf = cudf.read_csv(unsym_data, names=["src", "dst"], delimiter='\t', dtype=["int32", "int32"] )
# load the full symmetrized dataset for comparison
datafile='../data/karate-data.csv'
test_gdf = cudf.read_csv(datafile, names=["src", "dst"], delimiter='\t', dtype=["int32", "int32"] )
print("Unsymmetrized Graph")
print("\tNumber of Edges: " + str(len(gdf)))
print("Baseline Graph")
print("\tNumber of Edges: " + str(len(test_gdf)))
G = cugraph.Graph()
G.from_cudf_edgelist(gdf, source='src', destination='dst')
gdf_page = cugraph.pagerank(G)
# best PR score is
m = gdf_page['pagerank'].max()
df = gdf_page.query('pagerank == @m')
df
sdf = cugraph.symmetrize_df(gdf, 'src', 'dst')
print("Unsymmetrized Graph")
print("\tNumber of Edges: " + str(len(gdf)))
print("Symmetrized Graph")
print("\tNumber of Edges: " + str(len(sdf)))
print("Baseline Graph")
print("\tNumber of Edges: " + str(len(test_gdf)))
| 0.303835 | 0.910227 |
# One Shot Learning with Siamese Networks
This is the jupyter notebook that accompanies
## Imports
All the imports are defined here
```
%matplotlib inline
import torch
import numpy as np
from torch import optim
import torchvision.utils
import torch.nn.functional as F
from torch.autograd import Variable
import torchvision.transforms as transforms
from torch.utils.data import DataLoader,random_split
import config
from utils import imshow
from models import SiameseNetwork
from training import trainSiamese,inferenceSiamese
from datasets import SiameseNetworkDataset
from loss_functions import ContrastiveLoss
# generate_csv(config.training_dir)
import os
if not os.path.exists('state_dict'):
os.makedirs('state_dict')
```
## Using Image Folder Dataset
```
siamese_dataset = SiameseNetworkDataset(config.siamese_training_csv,
transform=transforms.Compose([
transforms.Resize((config.img_height,config.img_width)),
transforms.ToTensor(),
transforms.Normalize(0,1)]),
should_invert=False)
```
## Visualising some of the data
The top row and the bottom row of any column is one pair. The 0s and 1s correspond to the column of the image.
1 indiciates dissimilar, and 0 indicates similar.
```
# vis_dataloader = DataLoader(siamese_dataset,
# shuffle=True,
# num_workers=1,
# batch_size=8)
# dataiter = iter(vis_dataloader)
# example_batch = next(dataiter)
# concatenated = torch.cat((example_batch[0],example_batch[1]),0)
# imshow(torchvision.utils.make_grid(concatenated))
# print(example_batch[2].numpy())
```
## Training Time!
```
# Split the dataset into train, validation and test sets
num_train = round(0.9*siamese_dataset.__len__())
num_validate = siamese_dataset.__len__()-num_train
siamese_train, siamese_valid = random_split(siamese_dataset, [num_train,num_validate])
train_dataloader = DataLoader(siamese_train,
shuffle=True,
num_workers=8,
batch_size=config.train_batch_size)
valid_dataloader = DataLoader(siamese_valid,
shuffle=True,
num_workers=8,
batch_size=1)
net = SiameseNetwork().cuda()
criterion = ContrastiveLoss()
optimizer = optim.Adam(net.parameters(),lr = config.learning_rate )
scheduler = torch.optim.lr_scheduler.StepLR(optimizer,config.step_size, config.gamma)
net, train_loss_history, valid_loss_history,dict_name = trainSiamese(net,criterion,optimizer,scheduler,train_dataloader,
valid_dataloader,config.train_number_epochs,do_show=True)
```
## Testing
```
net = SiameseNetwork().cuda()
net.load_state_dict(torch.load(os.path.join("state_dict",dict_name)))
net.eval()
siamese_test = SiameseNetworkDataset(config.siamese_testing_csv,
transform=transforms.Compose([transforms.Resize((config.img_height,config.img_width)),
transforms.ToTensor(),
transforms.Normalize(0,1)
])
,should_invert=False)
test_dataloader = DataLoader(siamese_test,num_workers=8,batch_size=1,shuffle=True)
dataiter = iter(test_dataloader)
test_loss, test_er = inferenceSiamese(net,criterion,test_dataloader)
print("Test loss: %.4f\t Test error: %.4f"
%(test_loss, test_er))
for i in range(3):
label = 0
while label == 0:
x0,x1,label = next(dataiter)
label = label.detach().cpu().numpy()[0][0]
concatenated = torch.cat((x0,x1),0)
output1,output2 = net(Variable(x0).cuda(),Variable(x1).cuda())
euclidean_distance = F.pairwise_distance(output1, output2)
imshow(torchvision.utils.make_grid(concatenated),'Dissimilarity: {:.2f}\nLabel: {}'.format(euclidean_distance.item(),'Different'))
for i in range(3):
label = 1
while label == 1:
x0,x1,label = next(dataiter)
label = label.detach().cpu().numpy()[0][0]
concatenated = torch.cat((x0,x1),0)
output1,output2 = net(Variable(x0).cuda(),Variable(x1).cuda())
euclidean_distance = F.pairwise_distance(output1, output2)
imshow(torchvision.utils.make_grid(concatenated),'Dissimilarity: {:.2f}\nLabel: {}'.format(euclidean_distance.item(),'Same'))
dict_name
```
|
github_jupyter
|
%matplotlib inline
import torch
import numpy as np
from torch import optim
import torchvision.utils
import torch.nn.functional as F
from torch.autograd import Variable
import torchvision.transforms as transforms
from torch.utils.data import DataLoader,random_split
import config
from utils import imshow
from models import SiameseNetwork
from training import trainSiamese,inferenceSiamese
from datasets import SiameseNetworkDataset
from loss_functions import ContrastiveLoss
# generate_csv(config.training_dir)
import os
if not os.path.exists('state_dict'):
os.makedirs('state_dict')
siamese_dataset = SiameseNetworkDataset(config.siamese_training_csv,
transform=transforms.Compose([
transforms.Resize((config.img_height,config.img_width)),
transforms.ToTensor(),
transforms.Normalize(0,1)]),
should_invert=False)
# vis_dataloader = DataLoader(siamese_dataset,
# shuffle=True,
# num_workers=1,
# batch_size=8)
# dataiter = iter(vis_dataloader)
# example_batch = next(dataiter)
# concatenated = torch.cat((example_batch[0],example_batch[1]),0)
# imshow(torchvision.utils.make_grid(concatenated))
# print(example_batch[2].numpy())
# Split the dataset into train, validation and test sets
num_train = round(0.9*siamese_dataset.__len__())
num_validate = siamese_dataset.__len__()-num_train
siamese_train, siamese_valid = random_split(siamese_dataset, [num_train,num_validate])
train_dataloader = DataLoader(siamese_train,
shuffle=True,
num_workers=8,
batch_size=config.train_batch_size)
valid_dataloader = DataLoader(siamese_valid,
shuffle=True,
num_workers=8,
batch_size=1)
net = SiameseNetwork().cuda()
criterion = ContrastiveLoss()
optimizer = optim.Adam(net.parameters(),lr = config.learning_rate )
scheduler = torch.optim.lr_scheduler.StepLR(optimizer,config.step_size, config.gamma)
net, train_loss_history, valid_loss_history,dict_name = trainSiamese(net,criterion,optimizer,scheduler,train_dataloader,
valid_dataloader,config.train_number_epochs,do_show=True)
net = SiameseNetwork().cuda()
net.load_state_dict(torch.load(os.path.join("state_dict",dict_name)))
net.eval()
siamese_test = SiameseNetworkDataset(config.siamese_testing_csv,
transform=transforms.Compose([transforms.Resize((config.img_height,config.img_width)),
transforms.ToTensor(),
transforms.Normalize(0,1)
])
,should_invert=False)
test_dataloader = DataLoader(siamese_test,num_workers=8,batch_size=1,shuffle=True)
dataiter = iter(test_dataloader)
test_loss, test_er = inferenceSiamese(net,criterion,test_dataloader)
print("Test loss: %.4f\t Test error: %.4f"
%(test_loss, test_er))
for i in range(3):
label = 0
while label == 0:
x0,x1,label = next(dataiter)
label = label.detach().cpu().numpy()[0][0]
concatenated = torch.cat((x0,x1),0)
output1,output2 = net(Variable(x0).cuda(),Variable(x1).cuda())
euclidean_distance = F.pairwise_distance(output1, output2)
imshow(torchvision.utils.make_grid(concatenated),'Dissimilarity: {:.2f}\nLabel: {}'.format(euclidean_distance.item(),'Different'))
for i in range(3):
label = 1
while label == 1:
x0,x1,label = next(dataiter)
label = label.detach().cpu().numpy()[0][0]
concatenated = torch.cat((x0,x1),0)
output1,output2 = net(Variable(x0).cuda(),Variable(x1).cuda())
euclidean_distance = F.pairwise_distance(output1, output2)
imshow(torchvision.utils.make_grid(concatenated),'Dissimilarity: {:.2f}\nLabel: {}'.format(euclidean_distance.item(),'Same'))
dict_name
| 0.570571 | 0.906363 |
# Data Scrubber
Load the CSV data from `../data/raw` and clean it for processing. Cleaned data will be stored in `../data/clean`.
TODO:
* Jan 2020: Still getting clearly erroneous time stamps that go backward in time. Need to filter these out.
* Jan 2020: I started receiving CSV data from Govee that contained extra columns. See `office_interior_export202001131740.csv` :(
* Jan 2020: I started receiving CSV data from Govee that contained non-physical humidity level changes. Need to filter these out.
* Mar 2020: I should also load the clean data and use that as the first filter for dates I shouldn't load. That way I stop loading all the old files. Probably reverse iterate over the glob.
* If I do this, give myself a rewrite-all option also
```
import os
import glob
import pandas as pd
import util
import datetime as dt
import numpy as np
# invalidate any data points with a skip larger than this threshold
invalid_delta = pd.Timedelta('15.0 minutes')
# bad_data = all_data[all_data.timestamp < invalid_prior_to]
# bad_data
cur_dir = os.getcwd()
exterior_files = glob.glob(cur_dir.replace('notebooks','data/raw/*exterior*'))
interior_files = glob.glob(cur_dir.replace('notebooks','data/raw/*interior*'))
govee_columns = ['Timestamp for sample frequency every 1 min',
'Temperature_Celsius',
'Relative_Humidity']
def filter_single_dataset(df:pd.DataFrame=None):
return df
def combine_no_time_duplicates(files_glob):
all_data = pd.DataFrame(columns=govee_columns)
for f in files_glob:
data = pd.read_csv(f, warn_bad_lines=True, error_bad_lines=False)
data.set_index(keys=govee_columns[0])
# convert the correct column to datetime
data[govee_columns[0]] = pd.to_datetime(data[govee_columns[0]], infer_datetime_format=True, errors='coerce')
data = filter_single_dataset(data)
all_data = pd.concat([all_data, data],
ignore_index=True,
join='inner',
copy=True,
sort=True)
all_data[govee_columns[1]] = pd.to_numeric(all_data[govee_columns[1]], errors='coerce')
all_data[govee_columns[2]] = pd.to_numeric(all_data[govee_columns[2]], errors='coerce')
return all_data
all_exterior_data = combine_no_time_duplicates(exterior_files)
all_exterior_data['location'] = util.Locations.EXTERIOR.value
# all_exterior_data.to_csv('../data/clean/exterior.data.csv')
all_interior_data = combine_no_time_duplicates(interior_files)
all_interior_data['location'] = util.Locations.INTERIOR.value
# all_interior_data.to_csv('../data/clean/interior.data.csv')
# combine into one
all_data = pd.concat([all_exterior_data, all_interior_data])
all_data.rename(columns={govee_columns[0]: "timestamp",
govee_columns[1]: str(govee_columns[1]).lower(),
govee_columns[2]: str(govee_columns[2]).lower()},
inplace=True)
```
I've noticed invalid data coming from `Govee` and this needs to be handled. First, get a view of the bad data. Anything prior to Nov 9, 2019 is before I owned the device so we know that data must be invalid.
What's up with this, `Govee`?
```
# invalidate any data points prior to a certain date/time
invalid_prior_to = dt.datetime(year=2019, month=11, day=9, hour=15, minute=0)
bad_data = all_data[all_data.timestamp < invalid_prior_to]
bad_data
```
Now apply the timestamp filter to remove anamalous data.
```
strip_prior_to = dt.datetime(year=2019, month=11, day=9, hour=15, minute=30)
all_data = all_data[all_data.timestamp>=strip_prior_to]
timestamp_diff_threshold = dt.timedelta(minutes=5.0)
# above_threshold_series = all_data[govee_columns[0]].diff() > timestamp_diff_threshold
# all_data.drop(above_threshold_series.index)
```
# Write Clean Data
```
all_data.to_csv('../data/clean/sensor_data.csv', index=False)
os.listdir('../data/clean/')
all_data.head()
all_data.tail()
```
|
github_jupyter
|
import os
import glob
import pandas as pd
import util
import datetime as dt
import numpy as np
# invalidate any data points with a skip larger than this threshold
invalid_delta = pd.Timedelta('15.0 minutes')
# bad_data = all_data[all_data.timestamp < invalid_prior_to]
# bad_data
cur_dir = os.getcwd()
exterior_files = glob.glob(cur_dir.replace('notebooks','data/raw/*exterior*'))
interior_files = glob.glob(cur_dir.replace('notebooks','data/raw/*interior*'))
govee_columns = ['Timestamp for sample frequency every 1 min',
'Temperature_Celsius',
'Relative_Humidity']
def filter_single_dataset(df:pd.DataFrame=None):
return df
def combine_no_time_duplicates(files_glob):
all_data = pd.DataFrame(columns=govee_columns)
for f in files_glob:
data = pd.read_csv(f, warn_bad_lines=True, error_bad_lines=False)
data.set_index(keys=govee_columns[0])
# convert the correct column to datetime
data[govee_columns[0]] = pd.to_datetime(data[govee_columns[0]], infer_datetime_format=True, errors='coerce')
data = filter_single_dataset(data)
all_data = pd.concat([all_data, data],
ignore_index=True,
join='inner',
copy=True,
sort=True)
all_data[govee_columns[1]] = pd.to_numeric(all_data[govee_columns[1]], errors='coerce')
all_data[govee_columns[2]] = pd.to_numeric(all_data[govee_columns[2]], errors='coerce')
return all_data
all_exterior_data = combine_no_time_duplicates(exterior_files)
all_exterior_data['location'] = util.Locations.EXTERIOR.value
# all_exterior_data.to_csv('../data/clean/exterior.data.csv')
all_interior_data = combine_no_time_duplicates(interior_files)
all_interior_data['location'] = util.Locations.INTERIOR.value
# all_interior_data.to_csv('../data/clean/interior.data.csv')
# combine into one
all_data = pd.concat([all_exterior_data, all_interior_data])
all_data.rename(columns={govee_columns[0]: "timestamp",
govee_columns[1]: str(govee_columns[1]).lower(),
govee_columns[2]: str(govee_columns[2]).lower()},
inplace=True)
# invalidate any data points prior to a certain date/time
invalid_prior_to = dt.datetime(year=2019, month=11, day=9, hour=15, minute=0)
bad_data = all_data[all_data.timestamp < invalid_prior_to]
bad_data
strip_prior_to = dt.datetime(year=2019, month=11, day=9, hour=15, minute=30)
all_data = all_data[all_data.timestamp>=strip_prior_to]
timestamp_diff_threshold = dt.timedelta(minutes=5.0)
# above_threshold_series = all_data[govee_columns[0]].diff() > timestamp_diff_threshold
# all_data.drop(above_threshold_series.index)
all_data.to_csv('../data/clean/sensor_data.csv', index=False)
os.listdir('../data/clean/')
all_data.head()
all_data.tail()
| 0.308607 | 0.838283 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.